Identification of SMURF1 as a possible target for 7q21.3-22.1 amplification detected in a pancreatic cancer cell line by in-house array-based comparative genomic hybridization - PubMed (original) (raw)
Identification of SMURF1 as a possible target for 7q21.3-22.1 amplification detected in a pancreatic cancer cell line by in-house array-based comparative genomic hybridization
Ayako Suzuki et al. Cancer Sci. 2008 May.
Abstract
Pancreatic cancer (PC) cell lines provide a useful starting point for the discovery and functional analysis of genes driving the genesis and progression of this lethal cancer. To increase our understanding of the gene copy number changes in pancreatic carcinomas and to identify key amplification and deletion targets, we applied genome-wide array-based comparative genomic hybridization using in-house array (MCG Cancer Array-800) to 24 PC cell lines. Overall, the analyses revealed high genomic complexity, with several copy number changes detected in each line. Homozygous deletions (log(2)ratio < -2) of eight genes (clones) were seen in 14 of the 24 cell lines, whereas high-level amplifications (log(2)ratio > 2) of 10 genes (clones) were detected in seven lines. Among them, we focused on high-level amplification at 7q22.1, because target genes for this alteration remain unknown. Through precise mapping of the altered region by fluorescence in situ hybridization, determination of the expression status of genes located within those regions, and functional analysis using knockdown of the gene expression or the ectopic overexpression approach in PC cell lines, as well as immunohistochemical analyses of candidates in primary tumors of PC, we successfully identified SMURF1 as having the greatest potential as a 7q21.3-22.1 amplification target. SMURF1 may work as a growth-promoting gene in PC through overexpression and might be a good candidate as a therapeutic target. Our results suggest that array-based comparative genomic hybridization analysis combined with further genetic and functional examinations is a useful approach for identifying novel tumor-associated genes involved in the pathogenesis of this lethal disease.
Figures
Figure 1
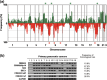
(a) Copy number analysis of pancreatic cancer (PC) cell lines using array‐based comparative genomic hybridization (array‐CGH) and (b) confirmation of homozygously deleted regions in primary tumors of PC. (a) Genome‐wide frequencies of copy number gains (above 0, green) and losses (below 0, red) in 24 PC cell lines. Clones are ordered from chromosomes 1–22, X, and Y, and within each chromosome on the basis of the UCSC mapping position (
, version May, 2004). Green asterisks, clones with at least one high‐level amplification; red asterisks, clones with at least one homozygous deletion. (b) Representative images from genomic polymerase chain reaction experiments showing glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), the functional control, and SMAD4, p16, N33, DEC1, and CDH23 exons 7 and 67 in 19 laser‐captured microdissection (LCM)‐treated primary PC. The frequency of homozygous deletion in each suppressor gene is shown.
Figure 2
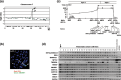
Amplification at 7q21.3‐22.1 in pancreatic cancer (PC) cell line. (a) Representative copy number profiles of chromosome 7 of AsPC‐1 cells in which array‐based comparative genomic hybridization (array‐CGH) analysis identified high‐level amplifications of SMURF1, TRRAP, and PDAP1 at 7q22.1 (arrowheads). Blue closed circles indicate genes (clones) showing a normal copy number ratio (–0.4 ≤ log2ratio ≤ 0.4), whereas green closed circles indicate those showing an increased copy number ratio (log2ratio > 0.4). (b) Representative fluorescence in situ hybridization (FISH) images with a bacterial artificial chromosome (BAC) clone containing the SMUFF1 gene (green signal, arrows and arrowheads) and a control BAC clone containing the CD6 gene (red signals, arrows) hybridized to metaphase chromosomes from AsPC‐1 cells, which showed a remarkably increased copy number of SMURF1 (arrow heads) with a homogeneously staining region (HSR) pattern. (c) Amplicon map of 7q21.3‐22.1 in the AsPC‐1 cell line. BAC used for FISH and their copy number are indicated as open bars and circles, respectively: those with >20 copies within the peak of HSR and outside of HSR in AsPC‐1 cells are shown in closed and open circles, respectively. Eleven genes located within the peak of amplification (closed arrow) showing an HSR pattern in AsPC‐1 cells are indicated as bars. (d) Expression of 11 genes located within the 7q21.3‐22.1 amplicon in 24 PC cell lines determined by reverse transcription–polymerase chain reaction. 1, AsPC‐1; 2, BxPC‐3; 3, Capan‐1; 4, Capan‐2; 5, CFPAC‐1; 6, HPAF‐II; 7, Hs766T; 8, KMP2; 9, KMP3; 10, KMP4; 11, KMP5; 12, KMP7; 13, KMP8; 14, KP1N; 15, KP1NL; 16, KP2; 17, KP3; 18, KP3L; 19, KP4‐4; 20, MIA‐Paca‐2; 21, PANC‐1; 22, PSN1; 23, SU.86.86; and 24, SW1990. Note that four genes, TRRAP, SMURF1, ARPC1A, and ARPC1B, showed remarkably increased expression in AsPC‐1 cells (arrow), and increased expression in several other cell lines compared to normal pancreas and normal pancreatic ductal cell‐derived line HPDE6.
Figure 3
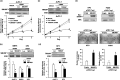
Growth‐promoting effects of SMURF1 and TRRAP1 on pancreatic cancer (PC) cell lines. (a) Effect of _SMURF1_‐small interfering RNA (siRNA) on growth of PC cell lines. AsPC‐1 cells were treated with 50 nM siRNA for SMURF1 (_SMURF1_‐siRNA) or control luciferase (_Luc_‐siRNA), or transfection reagent alone (mock). Upper, expression levels of SMURF1 protein 3 days after transfection were determined by western blotting using a specific antibody. Lower, relative cell number was determined by water‐soluble tetrazolium salt (WST) assay at the indicated times after transfection. Relative absorbance levels were calculated against the absorbance of cells before transfection (day 0). Data are the means ± SD of three separate experiments, each carried out in triplicate. Statistical analysis used the Mann–Whitney _U_‐test: (a) _SMURF_‐siRNA versus _Luc_‐siRNA, P < 0.05. (b) KMP4 and KP3 cell lines were treated with 50 nM _SMURF1_‐siRNA or control _Luc‐_siRNA, and analyses were carried out as described in (a). Relative absorbance levels of these cell lines 5 days after transfection are shown with the result of AsPC‐1 cell line. (c) Effect of _TRRAP_‐siRNA on the number of AsPC‐1 cells. AsPC‐1 cells were treated with 50 nM siRNA for _TRRAP_ (_TRRAP_‐siRNA) or control _luciferase_ (_Luc_‐siRNA), or transfection reagent alone (mock). Analyses were carried out as described in (a). (d) KP3 cells were treated with 50 nM _TRRAP_‐siRNA or control _Luc‐_siRNA, and analyses were carried out as described in (a). Relative absorbance levels in KP3 cell line 5 days after transfection are shown with the result of AsPC‐1 cell line. (e) Colony‐formation assay after transient ectopic expression of SMURF1 protein in KP3 and PSN1 cell lines. Myc‐tagged constructs containing the full coding sequence of _SMURF1_ (pCMV‐Tag3‐_SMURF1_) or empty vector (pCMV‐Tag3‐mock) as a control were transfected into cells, which showed a relatively low expression level of _SMURF1_. Upper, western blot analysis was carried out using 10 µg of protein extract and anti‐Myc tag antibody 48 h after transfection. Lower, 2 weeks after transfection of expression construct and subsequent selection of drug‐resistant colonies with appropriate concentrations of G418 in six‐well plates, the colonies formed were stained. Colonies >2 mm were counted, and the results are presented as the mean ± SD of three separate experiments, each carried out in triplicate. Statistical analysis used the Mann–Whitney U_‐test: (a) pCMV‐Tag3‐_SMURF1 versus pCMV‐Tag3‐mock, P < 0.05.
Figure 4
Expression of SMURF1 and TRRAP1 in primary pancreatic cancer (PC). Representative images of positive (left) and negative (right) immunohistochemical staining of SMURF1 (upper) and TRRAP (lower) proteins in primary tumors of PC (×100).
Similar articles
- SMURF1 amplification promotes invasiveness in pancreatic cancer.
Kwei KA, Shain AH, Bair R, Montgomery K, Karikari CA, van de Rijn M, Hidalgo M, Maitra A, Bashyam MD, Pollack JR. Kwei KA, et al. PLoS One. 2011;6(8):e23924. doi: 10.1371/journal.pone.0023924. Epub 2011 Aug 22. PLoS One. 2011. PMID: 21887346 Free PMC article. - Genome-wide array-based comparative genomic hybridization reveals multiple amplification targets and novel homozygous deletions in pancreatic carcinoma cell lines.
Heidenblad M, Schoenmakers EF, Jonson T, Gorunova L, Veltman JA, van Kessel AG, Höglund M. Heidenblad M, et al. Cancer Res. 2004 May 1;64(9):3052-9. doi: 10.1158/0008-5472.can-03-3159. Cancer Res. 2004. PMID: 15126341 - Detailed genomic mapping and expression analyses of 12p amplifications in pancreatic carcinomas reveal a 3.5-Mb target region for amplification.
Heidenblad M, Jonson T, Mahlamäki EH, Gorunova L, Karhu R, Johansson B, Höglund M. Heidenblad M, et al. Genes Chromosomes Cancer. 2002 Jun;34(2):211-23. doi: 10.1002/gcc.10063. Genes Chromosomes Cancer. 2002. PMID: 11979555 - Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer.
Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Bashyam MD, et al. Neoplasia. 2005 Jun;7(6):556-62. doi: 10.1593/neo.04586. Neoplasia. 2005. PMID: 16036106 Free PMC article. - Genome-wide analysis of pancreatic cancer using microarray-based techniques.
Harada T, Chelala C, Crnogorac-Jurcevic T, Lemoine NR. Harada T, et al. Pancreatology. 2009;9(1-2):13-24. doi: 10.1159/000178871. Epub 2008 Dec 12. Pancreatology. 2009. PMID: 19077451 Review.
Cited by
- Uncovering new insights into the role of the ubiquitin ligase Smurf1 on the regulation of innate immune signaling and resistance to infection.
Souza-Costa LP, Andrade-Chaves JT, Andrade JM, Costa VV, Franco LH. Souza-Costa LP, et al. Front Immunol. 2023 May 9;14:1185741. doi: 10.3389/fimmu.2023.1185741. eCollection 2023. Front Immunol. 2023. PMID: 37228615 Free PMC article. Review. - Expression profile of microRNAs in c-Myc induced mouse mammary tumors.
Sun Y, Wu J, Wu SH, Thakur A, Bollig A, Huang Y, Liao DJ. Sun Y, et al. Breast Cancer Res Treat. 2009 Nov;118(1):185-96. doi: 10.1007/s10549-008-0171-6. Epub 2008 Sep 7. Breast Cancer Res Treat. 2009. PMID: 18777135 Free PMC article. - miR-509-5p and miR-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer.
Hiramoto H, Muramatsu T, Ichikawa D, Tanimoto K, Yasukawa S, Otsuji E, Inazawa J. Hiramoto H, et al. Sci Rep. 2017 Jun 21;7(1):4002. doi: 10.1038/s41598-017-04191-w. Sci Rep. 2017. PMID: 28638102 Free PMC article. - SMURF1 amplification promotes invasiveness in pancreatic cancer.
Kwei KA, Shain AH, Bair R, Montgomery K, Karikari CA, van de Rijn M, Hidalgo M, Maitra A, Bashyam MD, Pollack JR. Kwei KA, et al. PLoS One. 2011;6(8):e23924. doi: 10.1371/journal.pone.0023924. Epub 2011 Aug 22. PLoS One. 2011. PMID: 21887346 Free PMC article. - Smad ubiquitylation regulatory factor 1/2 (Smurf1/2) promotes p53 degradation by stabilizing the E3 ligase MDM2.
Nie J, Xie P, Liu L, Xing G, Chang Z, Yin Y, Tian C, He F, Zhang L. Nie J, et al. J Biol Chem. 2010 Jul 23;285(30):22818-30. doi: 10.1074/jbc.M110.126920. Epub 2010 May 18. J Biol Chem. 2010. PMID: 20484049 Free PMC article.
References
- Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer 2002; 2: 897–909. - PubMed
- Matsuno S, Egawa S, Fukuyama S et al . Pancreatic cancer registry in Japan: 20 years of experience. Pancreas 2004; 28: 219–30. - PubMed
- Murr MM, Sarr MG, Oishi AJ, Van Heereden JA. Pancreatic cancer. CA Cancer J Clin 1994; 44: 304–18. - PubMed
- Grunewald K, Lyons J, Frohlich A et al . High frequency of Ki‐ras codon 12 mutations in pancreatic adenocarcinomas. Int J Cancer 1989; 43: 1037–41. - PubMed
- Moore PS, Sipos B, Orlandini S et al . Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K‐ras, p53, p16 and DPC4/Smad4. Virchows Arch 2001; 439: 798–802. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials