Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration - PubMed (original) (raw)
Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration
Jan Schmoranzer et al. Curr Biol. 2009.
Abstract
Background: Centrosome orientation toward the leading edge of migrating cells depends on dynein and microtubules (MTs), as well as a number of signaling factors at the leading edge. However, centrosomes are maintained at the cell center during orientation in fibroblasts, suggesting that factors working at sites other than the leading edge may also be involved.
Results: In a search for factors that function with dynein in centrosome orientation, we found that the polarity protein Par3 associated with dynein and that knockdown of Par3 inhibited centrosome orientation by disrupting the position of the centrosome at the cell center; this disrupted centrosome positioning is the same phenotype as that observed with dynein inhibition. Par3 associated with dynein through its N-terminal dimerization and PDZ1 domains and interacted specifically with dynein light intermediate chain 2 (LIC2). siRNA knockdown of LIC2, but not LIC1, or overexpression of LIC2 or the N-terminal domain of Par3, also inhibited centrosome orientation by disrupting centrosome position. In wound-edge fibroblasts, Par3 specifically localized to cell-cell contacts where it overlapped with MT ends and dynein puncta in a LIC2-dependent fashion. Live imaging showed that MTs exhibited increased pausing at cell-cell contacts compared to the leading edge and that this elevated pausing was dependent on Par3 and LIC2.
Conclusions: Par3 associates with dynein and contributes to the local regulation of MT dynamics at cell-cell contacts and proper positioning of the centrosome at the cell center. We propose that Par3 acts as a cortical factor that tethers MTs through its association with LIC2 dynein.
Figures
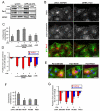
Figure 1. Par3 association with dynein
(A) Dynein immunoprecipitation from lysates of NIH3T3 cells using αDIC or control mouse IgG (MIgG), western blotted for endogenous Par3, Par6, PKCζ, Lgl-1, β-catenin (β-cat) and the p150_Glued_ subunit of dynactin. Immunoprecipitation of dynein was confirmed by western blot for DIC and DHC. Both isoforms of Par3 (180kDa stronger than 100kDa) coimmunoprecipitated with dynein. (B) YFP-Par3 immunoprecipitation from lysates of COS-7 cells expressing YFP-Par3 using GFP antibody that recognizes YFP (αYFP) or control rabbit IgG (RbIgG), western blotted for DIC or YFP-Par3. (C) Dynein immunoprecipitation from COS-7 cells expressing YFP-Par3 full length (Par3) or fragments as indicated using αDIC or control mouse IgG (MIgG), western blotted for YFP-Par3 and DIC. (D) YFP-Par3 immunoprecipitation from COS-7 cells co-expressing YFP-Par3 and Flag-, Myc- or HA-tagged subunits of dynein using GFP antibody (labeled αYFP in figure) or control rabbit IgG (RbIgG). Western blots for dynein subunit tags or YFP-Par3. In A-D, “Input” was 2-4% of total cell lysates.
Figure 2. Par3 is necessary for centrosome orientation and centration
(A) Western blots of Par3 and mDia1 (loading control) from NIH3T3 cells (wt and a stable cell line expressing GFP-α-tubulin) treated with siRNAs to Par3 and GAPDH (control). (B) Representative images of GAPDH and Par3 siRNA-treated NIH3T3 cells that were wounded, LPA-stimulated and stained for Par3, β-catenin and pericentrin (in one channel), and MTs (shown as an overlay with Par3). Nuclear staining was not reduced by Par3 siRNA suggesting that it is a staining artifact of the commercial antibody. Bar, 20μm. (C) Quantification of LPA-stimulated centrosome orientation in GAPDH or Par3 siRNA-treated and αDIC injected cells. Centrosomes were scored as oriented when they were positioned in the triangular sector defined by the nucleus and leading edge [2]. Random centrosome orientation is 33% with these criteria. (D) Quantification of the position of the nucleus (red) and the centrosome (blue) along the axis of the cell perpendicular to the wound in LPA-stimulated GAPDH and Par3 siRNA-treated cells and αDIC injected cells. The cell center is defined as “0”; (+) values are toward the leading edge and (-) are toward the rear of the cell. Values are plotted as percentage of cell radius to normalize for differences in cell area. (E) Representative images of wounded and LPA-stimulated cells expressing a control YFP-Par3 fragment (205-208) and dynein-binding YFP-Par3 fragments (Nt-90, PDZ1). Cells were stained for MTs (green), Par3 tag (red) and DAPI (blue). Bar, 20μm. (F) Quantification of LPA-stimulated centrosome orientation in cells expressing Par3 fragments. (G) Quantification of the position of the nucleus (red) and the centrosome (blue) as in (D) in LPA-stimulated cells expressing Par3 fragments as indicated. Centrosome orientation data (C,F) and nuclear and centrosome position data (D,G) are based upon at least two separate experiments (N>50 cells for each condition). C,F: error bars = s.d. (N>50 cells for each condition). D,G: error bars = s.e.m.
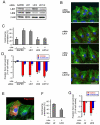
Figure 3. Dynein LIC2 is necessary for centrosome orientation and centration
(A) Western blots of LIC1, LIC2 and β-catenin (100 kDa, loading control) from NIH3T3 cells treated with the indicated siRNAs. See Figure S2 for quantification. (B) Representative images of wounded and LPA-stimulated NIH3T3 cells treated with the indicated siRNA. Cells were stained for MTs (green), N-cadherin + pericentrin (red) and DAPI (blue). The centrosome appears as a yellow spot due to overlap of pericentrin and MT staining. Bar, 20μm. (C) Quantification of LPA-stimulated centrosome orientation in GAPDH, LIC1 and LIC2 depleted cells. (D) Quantification of the position of the nucleus (red) and the centrosome (blue) (as defined in legend of Figure 2D) in LPA-stimulated cells depleted of LIC1, LIC2 or GAPDH (control). . (E) Representative image of a wounded and LPA-stimulated NIH3T3 cell expressing Flag-LIC2. Cells were stained for MTs (green), LIC2 tag (red) and DAPI (blue). Bar, 20μm. (F) Quantification of LPA-stimulated centrosome orientation in control GFP (ctr) and Flag-LIC2 expressing NIH3T3 cells. (G) Quantification of the position of the nucleus (red) and the centrosome (blue) (as defined in legend of Figure 2D) in LPA-stimulated cells expressing control GFP (ctr) or LIC2 as indicated. Centrosome orientation (C,F) and nuclear/centrosome positioning data (D,G) are based upon at least two separate experiments (N>50 cells for each condition). Error bars = s.e.m.
Figure 4. Par3 depletion and dominant negative construct interfere with cell migration
(A) Images from movies of wounded monolayers of NIH3T3 cells that had been treated with siRNA against GAPDH or Par3 and allowed to migrate in the presence of serum. Wound edge is outlined in black. Bar, 100μm. (B) Frequency histogram of individual wound edge speeds (binned in 1μm/h intervals) and the average migration speeds (inset histogram) of control GAPDH and Par3 siRNA-treated cells. Data represents 60 wound edges and two independent experiments. Error bars = s.d. Statistical significance was assessed with a two-tailed, unpaired T-test (*** = p<0.0001). **(C)** Images of NIH3T3 cells expressing YFP-Par3 205-283 or YFP-Par3 Nt-90 and stimulated to migrate for 15 hr with 2% serum. Cells were stained for GFP (red), MTs (green) and DAPI (blue). Constructs were expressed in wound-edge cells by cDNA microinjection just after wounding. Bar, 20μm. **(D)** Quantification of the wound edge persistence of NIH3T3 cells expressing the indicated Par3 fragments after 15 hr migration in 2% serum. Data are from at least four separate experiments with a total number of injected cells > 50 (error bars = s.d.). Statistical significance was assessed with Chi2-test (*** = p<0.0001).
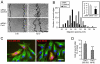
Figure 5. Par3 localizes to cell-cell contacts of wound-edge NIH3T3 cells where it overlaps with MT ends and dynein
(A) Untreated and (C) LPA-stimulated woundedge NIH3T3 cells stained for MTs (green) and endogenous Par3 (red). Bar, 20μm. (B,D) High magnification of boxed regions labeled “1” and “2” in A and C. Bar, 5 μm. Arrows indicate puncta of Par3 that colocalize with MT ends. (E) Quantification of Par3 overlap with MTs at cell-cell contacts in cells treated with the indicated siRNAs and stimulated with LPA. (F,G) Wounded and LPA-stimulated NIH3T3 cells stained for DIC and Par3. (F) Low magnification view showing punctate DIC distribution throughout the cytosol with accumulations in the perinuclear area and the leading edge. Bar, 20μm. (G) Higher magnification of boxed region in F. Arrows indicate overlapping puncta of DIC and Par3. Bar, 5μm. (H) Quantification of Par3 overlap with DIC at cell-cell contacts in cells treated with the indicated siRNAs and stimulated with LPA. Data (E,H) are from two independent experiments in which more than 100 regions from 10-15 images were analyzed. Error bars = s.e.m. Statistical significance was assessed with a two-tailed, unpaired T-test (*** = p<0.0001; ns =not significant).
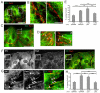
Figure 6. LPA stimulates Par3 and LIC2 dependent pausing of MTs at the cell-cell contacts
(A) Frames from movies of GFP-α-tubulin expressing NIH3T3 cells showing examples of MT dynamics at regions of cell-cell contact before (-LPA) and after stimulation with LPA (+LPA). The first panel shows a phase image from the beginning of the movie (dashed line denotes cell-cell contact). Arrowheads indicate MTs that exhibit dynamic instability in unstimulated cells (-LPA) and a MT that pauses for 84 sec (frames 14-98s) in an LPA-stimulated cell (+LPA). Bars, 2μm. (B) Life history plots (distance vs. time) for 3 MT ends at the leading edge and cell-cell contacts of GFP-α-tubulin expressing NIH3T3 cells before (-LPA) and after stimulation with LPA (+LPA). (C) Quantification of time MTs spent pausing and (D) MTs that exhibited long pause intervals (>30 sec) at cell-cell contacts and the leading edge from GFP-α-tubulin expressing NIH3T3 cells treated as indicated. Data for each condition are pooled from 4-8 different cells (2-3 independent experiments) and 25-47 MTs. (E) Quantification of time MTs spent pausing and (F) MTs that exhibited long pause intervals (>30 sec) at cell-cell contacts of Cy3-tubulin injected cells expressing YFP-Par3 205-283 (ctr) and Nt-90 fragments. Data for each condition are from 6-10 different cells (2 independent experiments) and 23-46 total MTs. In C-F, error bars are the 95% confidence intervals. Statistical significance was assessed with Fisher exact test (* = p<0.05, ** = p<0.001, ***=p<0.0001, “ns” is not significant).
Similar articles
- POPX2 phosphatase regulates cell polarity and centrosome placement.
Hoon JL, Li HY, Koh CG. Hoon JL, et al. Cell Cycle. 2014;13(15):2459-68. doi: 10.4161/cc.29421. Cell Cycle. 2014. PMID: 25483195 Free PMC article. - Cytoplasmic dynein is involved in the retention of microtubules at the centrosome in interphase cells.
Burakov A, Kovalenko O, Semenova I, Zhapparova O, Nadezhdina E, Rodionov V. Burakov A, et al. Traffic. 2008 Apr;9(4):472-80. doi: 10.1111/j.1600-0854.2007.00698.x. Epub 2007 Dec 26. Traffic. 2008. PMID: 18182007 - FAK regulates dynein localisation and cell polarity in migrating mouse fibroblasts.
Fructuoso M, Legrand M, Mousson A, Steffan T, Vauchelles R, De Mey J, Sick E, Rondé P, Dujardin D. Fructuoso M, et al. Biol Cell. 2020 Feb;112(2):53-72. doi: 10.1111/boc.201900041. Epub 2020 Jan 9. Biol Cell. 2020. PMID: 31859373 - [Dynein and dynactin as organizers of the system of cell microtubules].
Burakov AV, Nadezhdina ES. Burakov AV, et al. Ontogenez. 2006 Sep-Oct;37(5):323-39. Ontogenez. 2006. PMID: 17066975 Review. Russian. - End-on microtubule-dynein interactions and pulling-based positioning of microtubule organizing centers.
Laan L, Roth S, Dogterom M. Laan L, et al. Cell Cycle. 2012 Oct 15;11(20):3750-7. doi: 10.4161/cc.21753. Epub 2012 Aug 16. Cell Cycle. 2012. PMID: 22895049 Free PMC article. Review.
Cited by
- Centriole Translational Planar Polarity in Monociliated Epithelia.
Donati A, Schneider-Maunoury S, Vesque C. Donati A, et al. Cells. 2024 Aug 23;13(17):1403. doi: 10.3390/cells13171403. Cells. 2024. PMID: 39272975 Free PMC article. Review. - CCSer2 gates dynein activity at the cell periphery.
Zang JL, Gibson D, Zheng AM, Shi W, Gillies JP, Stein C, Drerup CM, DeSantis ME. Zang JL, et al. bioRxiv [Preprint]. 2024 Jun 14:2024.06.13.598865. doi: 10.1101/2024.06.13.598865. bioRxiv. 2024. PMID: 38915497 Free PMC article. Preprint. - The receptor protein tyrosine phosphatase PTPRK promotes intestinal repair and catalysis-independent tumour suppression.
Young KA, Wojdyla K, Lai T, Mulholland KE, Aldaz Casanova S, Antrobus R, Andrews SR, Biggins L, Mahler-Araujo B, Barton PR, Anderson KR, Fearnley GW, Sharpe HJ. Young KA, et al. J Cell Sci. 2024 Jul 15;137(14):jcs261914. doi: 10.1242/jcs.261914. Epub 2024 Jul 22. J Cell Sci. 2024. PMID: 38904097 Free PMC article. - Systemic cellular migration: The forces driving the directed locomotion movement of cells.
De la Fuente IM, Carrasco-Pujante J, Camino-Pontes B, Fedetz M, Bringas C, Pérez-Samartín A, Pérez-Yarza G, López JI, Malaina I, Cortes JM. De la Fuente IM, et al. PNAS Nexus. 2024 Apr 20;3(5):pgae171. doi: 10.1093/pnasnexus/pgae171. eCollection 2024 May. PNAS Nexus. 2024. PMID: 38706727 Free PMC article. - Regulation of Cell Adhesion and Migration via Microtubule Cytoskeleton Organization, Cell Polarity, and Phosphoinositide Signaling.
Thapa N, Wen T, Cryns VL, Anderson RA. Thapa N, et al. Biomolecules. 2023 Sep 22;13(10):1430. doi: 10.3390/biom13101430. Biomolecules. 2023. PMID: 37892112 Free PMC article. Review.
References
- Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. - PubMed
- Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, Gundersen GG. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11:1536–1541. - PubMed
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. - PubMed
- Manneville JB, Etienne-Manneville S. Positioning centrosomes and spindle poles: looking at the periphery to find the centre. Biol Cell. 2006;98:557–565. - PubMed
- Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nature reviews. 2008;9:860–873. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM047434/GM/NIGMS NIH HHS/United States
- R01 GM062939/GM/NIGMS NIH HHS/United States
- R01 GM062938/GM/NIGMS NIH HHS/United States
- GM47434/GM/NIGMS NIH HHS/United States
- R01 GM062939-06/GM/NIGMS NIH HHS/United States
- R01 GM047434/GM/NIGMS NIH HHS/United States
- R01 GM062939-07/GM/NIGMS NIH HHS/United States
- R01 GM062939-08/GM/NIGMS NIH HHS/United States
- GM062939/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases