A human gut microbial gene catalogue established by metagenomic sequencing - PubMed (original) (raw)
Comparative Study
. 2010 Mar 4;464(7285):59-65.
doi: 10.1038/nature08821.
Ruiqiang Li, Jeroen Raes, Manimozhiyan Arumugam, Kristoffer Solvsten Burgdorf, Chaysavanh Manichanh, Trine Nielsen, Nicolas Pons, Florence Levenez, Takuji Yamada, Daniel R Mende, Junhua Li, Junming Xu, Shaochuan Li, Dongfang Li, Jianjun Cao, Bo Wang, Huiqing Liang, Huisong Zheng, Yinlong Xie, Julien Tap, Patricia Lepage, Marcelo Bertalan, Jean-Michel Batto, Torben Hansen, Denis Le Paslier, Allan Linneberg, H Bjørn Nielsen, Eric Pelletier, Pierre Renault, Thomas Sicheritz-Ponten, Keith Turner, Hongmei Zhu, Chang Yu, Shengting Li, Min Jian, Yan Zhou, Yingrui Li, Xiuqing Zhang, Songgang Li, Nan Qin, Huanming Yang, Jian Wang, Søren Brunak, Joel Doré, Francisco Guarner, Karsten Kristiansen, Oluf Pedersen, Julian Parkhill, Jean Weissenbach; MetaHIT Consortium; Peer Bork, S Dusko Ehrlich, Jun Wang
Collaborators, Affiliations
- PMID: 20203603
- PMCID: PMC3779803
- DOI: 10.1038/nature08821
Comparative Study
A human gut microbial gene catalogue established by metagenomic sequencing
Junjie Qin et al. Nature. 2010.
Abstract
To understand the impact of gut microbes on human health and well-being it is crucial to assess their genetic potential. Here we describe the Illumina-based metagenomic sequencing, assembly and characterization of 3.3 million non-redundant microbial genes, derived from 576.7 gigabases of sequence, from faecal samples of 124 European individuals. The gene set, approximately 150 times larger than the human gene complement, contains an overwhelming majority of the prevalent (more frequent) microbial genes of the cohort and probably includes a large proportion of the prevalent human intestinal microbial genes. The genes are largely shared among individuals of the cohort. Over 99% of the genes are bacterial, indicating that the entire cohort harbours between 1,000 and 1,150 prevalent bacterial species and each individual at least 160 such species, which are also largely shared. We define and describe the minimal gut metagenome and the minimal gut bacterial genome in terms of functions present in all individuals and most bacteria, respectively.
Figures
Figure 1. Coverage of human gut microbiome
The three human microbial sequencing read sets, Illumina GA reads generated from 124 individuals in this study (black; n=124), Roche/454 reads from 18 human twins and their mothers (grey; n=18), and Sanger reads from 13 Japanese individuals (white; n=13) were aligned to each of the reference sequence sets. Mean values ± s.e.m. are plotted.
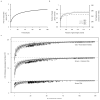
Figure 2. Predicted ORFs in the human gut microbiomes
a, Number of unique genes as function of the extent of sequencing. The gene accumulation curve corresponds to the Sobs (Mao Tau) values, calculated using EstimateS(version 8.2.0) on randomly chosen 100 samples (due to memory limitation). b, Coverage of genes from 89 frequent gut microbial species (Supplementary Table 12). c, Number of functions captured by number of samples investigated, based upon known (well characterized) orthologous groups (OGs; bottom), known+unknown orthologous groups (including e.g. putative, predicted, conserved hypothetical functions; center) and OGs+novel gene families (>20 proteins) recovered from the metagenome (top).
Figure 3. Relative abundance of frequent microbial genomes among individuals of the cohort
Boxes denote 25% and 75% percentiles, the black line in the box corresponds to the median, the “whiskers” indicate the interquartile range from either or both ends of the box, the dots show the outliers, beyond the ends of the whiskers (See supplementary Methods for computation).
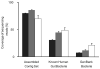
Figure 4. Bacterial species abundance differentiates IBD patients and healthy individuals
Principal component analysis based on the abundance of 155 species with ≥1% genome coverage by the Illumina reads in at least 1 individual of the cohort was carried out with 14 healthy individuals and 25 IBD patients from Spain.
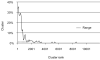
Figure 5. Clusters that contain the B. subtilis essential genes
The clusters were ranked by the number of genes they contain, normalized by average length and copy number (see Supplementary Fig. 10) and the proportion of clusters with the essential B. subtilis genes was determined for successive groups of 100 clusters. Range indicates the part of the cluster distribution that contains 86 % of the B. subtilis essential genes.
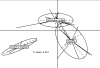
Figure 6. Characterization of the minimal gut genome and metagenome
a, Projection of the minimal gut genome on the KEGG pathways using the Ipath tool. b, Functional composition of the minimal gut genome and metagenome. c, Estimation of the minimal gut metagenome size. Known orthologous groups (OGs; red), known+unknown OGs (blue) and OGs+novel gene families (>20 proteins; grey). Inset: Composition of the gut minimal microbiome. Large circle: Classification in the minimal metagenome according to OG occurrence in STRING7 bacterial genomes. Common (25%), uncommon (35%) and rare (45%) are present in >50%, <50% but >10% and <10% of genomes, respectively. Small circle: composition of the rare OGs. Unknown (80%) have no annotation or are poorly characterized, while known bacterial (19%) and phage-related (1%) OGs have functional description.
Comment in
- A gut feeling for disease.
Flintoft L. Flintoft L. Nat Rev Genet. 2010 Apr;11(4):237. doi: 10.1038/nrg2767. Nat Rev Genet. 2010. PMID: 21488225 No abstract available.
Similar articles
- Mining biomass-degrading genes through Illumina-based de novo sequencing and metagenomic analysis of free-living bacteria in the gut of the lower termite Coptotermes gestroi harvested in Vietnam.
Do TH, Nguyen TT, Nguyen TN, Le QG, Nguyen C, Kimura K, Truong NH. Do TH, et al. J Biosci Bioeng. 2014 Dec;118(6):665-71. doi: 10.1016/j.jbiosc.2014.05.010. Epub 2014 Jun 11. J Biosci Bioeng. 2014. PMID: 24928651 - Binning metagenomic contigs by coverage and composition.
Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Alneberg J, et al. Nat Methods. 2014 Nov;11(11):1144-6. doi: 10.1038/nmeth.3103. Epub 2014 Sep 14. Nat Methods. 2014. PMID: 25218180 - [Metagenomics of the intestinal microbiota: potential applications].
Dusko Ehrlich S; MetaHIT consortium. Dusko Ehrlich S, et al. Gastroenterol Clin Biol. 2010 Sep;34 Suppl 1:S23-8. doi: 10.1016/S0399-8320(10)70017-8. Gastroenterol Clin Biol. 2010. PMID: 20889001 French. - Gastrointestinal microbiology enters the metagenomics era.
Frank DN, Pace NR. Frank DN, et al. Curr Opin Gastroenterol. 2008 Jan;24(1):4-10. doi: 10.1097/MOG.0b013e3282f2b0e8. Curr Opin Gastroenterol. 2008. PMID: 18043225 Review. - Diversity and genomes of uncultured microbial symbionts in the termite gut.
Hongoh Y. Hongoh Y. Biosci Biotechnol Biochem. 2010;74(6):1145-51. doi: 10.1271/bbb.100094. Epub 2010 Jun 7. Biosci Biotechnol Biochem. 2010. PMID: 20530908 Review.
Cited by
- Fecal Microbiota Transplantation: Indications, Methods, and Challenges.
Lee JY, Kim Y, Kim J, Kim JK. Lee JY, et al. J Microbiol. 2024 Nov 18. doi: 10.1007/s12275-024-00184-3. Online ahead of print. J Microbiol. 2024. PMID: 39557804 Review. - Functional Outcome Prediction of Acute Ischemic Stroke Based on the Oral and Gut Microbiota.
Liang J, Ren Y, Zheng Y, Lin X, Song W, Zhu J, Zhang X, Zhou H, Wu Q, He Y, Yin J. Liang J, et al. Mol Neurobiol. 2024 Nov 15. doi: 10.1007/s12035-024-04618-2. Online ahead of print. Mol Neurobiol. 2024. PMID: 39546118 - Influence of gender, age, and body mass index on the gut microbiota of individuals from South China.
Li S, Fan S, Ma Y, Xia C, Yan Q. Li S, et al. Front Cell Infect Microbiol. 2024 Oct 31;14:1419884. doi: 10.3389/fcimb.2024.1419884. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39544283 Free PMC article. - Reverse engineering the Gut-Brain Axis and microbiome-metabolomics for symbiotic/pathogenic balance in neurodegenerative diseases.
Munir MU, Ali SA, Chung KHK, Kakinen A, Javed I, Davis TP. Munir MU, et al. Gut Microbes. 2024 Jan-Dec;16(1):2422468. doi: 10.1080/19490976.2024.2422468. Epub 2024 Nov 10. Gut Microbes. 2024. PMID: 39523450 Free PMC article. Review. - Impact of Dietary Fatty Acid Composition on the Intestinal Microbiota and Fecal Metabolism of Rats Fed a High-Fructose/High-Fat Diet.
Zhao Z, Zhong L, Zhou P, Deng Y, Liu G, Li P, Zeng J, Zhang Y, Tang X, Zhang M. Zhao Z, et al. Nutrients. 2024 Nov 3;16(21):3774. doi: 10.3390/nu16213774. Nutrients. 2024. PMID: 39519607 Free PMC article.
References
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi:S0092-8674(06)00192-9 [pii]10.1016/j.cell.2006.02.017. - PubMed
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi:307/5717/1915 [pii]10.1126/science.1104816. - PubMed
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi:10.1146/annurev.nutr.22.011602.092259011602.092259 [pii] - PubMed
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi:4441022a [pii]10.1038/4441022a. - PubMed
- Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi:nature05414 [pii]10.1038/nature05414. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources