A MOD(ern) perspective on literature curation - PubMed (original) (raw)
A MOD(ern) perspective on literature curation
Jodi Hirschman et al. Mol Genet Genomics. 2010 May.
Abstract
Curation of biological data is a multi-faceted task whose goal is to create a structured, comprehensive, integrated, and accurate resource of current biological knowledge. These structured data facilitate the work of the scientific community by providing knowledge about genes or genomes and by generating validated connections between the data that yield new information and stimulate new research approaches. For the model organism databases (MODs), an important source of data is research publications. Every published paper containing experimental information about a particular model organism is a candidate for curation. All such papers are examined carefully by curators for relevant information. Here, four curators from different MODs describe the literature curation process and highlight approaches taken by the four MODs to address: (1) the decision process by which papers are selected, and (2) the identification and prioritization of the data contained in the paper. We will highlight some of the challenges that MOD biocurators face, and point to ways in which researchers and publishers can support the work of biocurators and the value of such support.
Figures
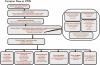
Fig. 1
A typical curation workflow, exemplified by the process at ZFIN. Curation workflows are unique as each MOD strives to best serve its own research community. For example at some MODS, different members of the curation team may enter different types of data, whereas at other MODS a single curator may enter all of the data types from a paper. Additional differences in workflow stem mainly from staffing and other budgetary constraints for each database. However, there are many commonalities in the workflow process, as the questions that must be answered to complete curation of a paper are similar regardless of the MOD. Here, the curation workflow at ZFIN illustrates the order in which certain tasks take place and many of the questions that must be answered at each step. Papers that lack key details can prevent curators from answering questions critical to the curation process, leading to a reduction in the amount or the detail of the curated data
Fig. 2
A TAIR web query form using controlled vocabularies to ask “Find a gene whose symbol begins with At1g and has GO function annotations based on direct assays, and codes for a protein that has literature associated with it.”
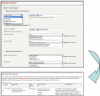
Fig. 3
Snapshot of the Gene Detail page for Fech (upper left), and snapshots of MGI editorial interfaces for input of data relating to symbol, name, and synonyms (upper right), phenotypic alleles (bottom right), including expanded window showing a controlled pick list, and mammalian homology (bottom left). Arrows point to the relevant section of the gene detail page that the editorial interface addresses
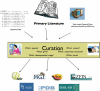
Fig. 4
The global flow of biological data, as presented from a MOD perspective. Curators read the published literature and data that can be extracted for the database is identified and entered. Other sources of data may also be incorporated and in some cases can be used to identify inconsistencies with the literature-derived data. The curation process serves to organize and integrate data into the relational database format for users to easily view what is known and not known about their favorite genes or proteins
Similar articles
- Directly e-mailing authors of newly published papers encourages community curation.
Bunt SM, Grumbling GB, Field HI, Marygold SJ, Brown NH, Millburn GH; FlyBase Consortium. Bunt SM, et al. Database (Oxford). 2012 May 2;2012:bas024. doi: 10.1093/database/bas024. Print 2012. Database (Oxford). 2012. PMID: 22554788 Free PMC article. - Biocuration at the Saccharomyces genome database.
Skrzypek MS, Nash RS. Skrzypek MS, et al. Genesis. 2015 Aug;53(8):450-7. doi: 10.1002/dvg.22862. Epub 2015 Jul 3. Genesis. 2015. PMID: 25997651 Free PMC article. Review. - Canto: an online tool for community literature curation.
Rutherford KM, Harris MA, Lock A, Oliver SG, Wood V. Rutherford KM, et al. Bioinformatics. 2014 Jun 15;30(12):1791-2. doi: 10.1093/bioinformatics/btu103. Epub 2014 Feb 25. Bioinformatics. 2014. PMID: 24574118 Free PMC article. - Automatic categorization of diverse experimental information in the bioscience literature.
Fang R, Schindelman G, Van Auken K, Fernandes J, Chen W, Wang X, Davis P, Tuli MA, Marygold SJ, Millburn G, Matthews B, Zhang H, Brown N, Gelbart WM, Sternberg PW. Fang R, et al. BMC Bioinformatics. 2012 Jan 26;13:16. doi: 10.1186/1471-2105-13-16. BMC Bioinformatics. 2012. PMID: 22280404 Free PMC article. - The curation of genetic variants: difficulties and possible solutions.
Pandey KR, Maden N, Poudel B, Pradhananga S, Sharma AK. Pandey KR, et al. Genomics Proteomics Bioinformatics. 2012 Dec;10(6):317-25. doi: 10.1016/j.gpb.2012.06.006. Epub 2012 Nov 29. Genomics Proteomics Bioinformatics. 2012. PMID: 23317699 Free PMC article. Review.
Cited by
- pubmed2ensembl: a resource for mining the biological literature on genes.
Baran J, Gerner M, Haeussler M, Nenadic G, Bergman CM. Baran J, et al. PLoS One. 2011;6(9):e24716. doi: 10.1371/journal.pone.0024716. Epub 2011 Sep 29. PLoS One. 2011. PMID: 21980353 Free PMC article. - Biocuration: Distilling data into knowledge.
International Society for Biocuration. International Society for Biocuration. PLoS Biol. 2018 Apr 16;16(4):e2002846. doi: 10.1371/journal.pbio.2002846. eCollection 2018 Apr. PLoS Biol. 2018. PMID: 29659566 Free PMC article. - Outreach and online training services at the Saccharomyces Genome Database.
MacPherson KA, Starr B, Wong ED, Dalusag KS, Hellerstedt ST, Lang OW, Nash RS, Skrzypek MS, Engel SR, Cherry JM. MacPherson KA, et al. Database (Oxford). 2017 Jan 1;2017(1):bax002. doi: 10.1093/database/bax002. Database (Oxford). 2017. PMID: 28365719 Free PMC article. - GermOnline 4.0 is a genomics gateway for germline development, meiosis and the mitotic cell cycle.
Lardenois A, Gattiker A, Collin O, Chalmel F, Primig M. Lardenois A, et al. Database (Oxford). 2010 Dec 11;2010:baq030. doi: 10.1093/database/baq030. Print 2010. Database (Oxford). 2010. PMID: 21149299 Free PMC article. - Text mining effectively scores and ranks the literature for improving chemical-gene-disease curation at the comparative toxicogenomics database.
Davis AP, Wiegers TC, Johnson RJ, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, Murphy CG, Mattingly CJ. Davis AP, et al. PLoS One. 2013 Apr 17;8(4):e58201. doi: 10.1371/journal.pone.0058201. Print 2013. PLoS One. 2013. PMID: 23613709 Free PMC article.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
- Ausdesirk T, Audesirk G, Byers B. Life on earth. 3. Saddlebrook: Pearson Prentice Hall; 2004.
Publication types
MeSH terms
Grants and funding
- P41 HG002659/HG/NHGRI NIH HHS/United States
- R01 GM080646/GM/NIGMS NIH HHS/United States
- P41 HG000330/HG/NHGRI NIH HHS/United States
- P41 HG001315/HG/NHGRI NIH HHS/United States
- GM080646/GM/NIGMS NIH HHS/United States
- P41 HG002273/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources