Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia - PubMed (original) (raw)
doi: 10.3324/haematol.2010.039768. Epub 2011 May 5.
Larry Mansouri, Anders Isaksson, Hanna Göransson, Nicola Cahill, Mattias Jansson, Markus Rasmussen, Jeanette Lundin, Stefan Norin, Anne Mette Buhl, Karin Ekström Smedby, Henrik Hjalgrim, Karin Karlsson, Jesper Jurlander, Christian Geisler, Gunnar Juliusson, Richard Rosenquist
Affiliations
- PMID: 21546498
- PMCID: PMC3148910
- DOI: 10.3324/haematol.2010.039768
Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia
Rebeqa Gunnarsson et al. Haematologica. 2011 Aug.
Abstract
Background: High-resolution genomic microarrays enable simultaneous detection of copy-number aberrations such as the known recurrent aberrations in chronic lymphocytic leukemia [del(11q), del(13q), del(17p) and trisomy 12], and copy-number neutral loss of heterozygosity. Moreover, comparison of genomic profiles from sequential patients' samples allows detection of clonal evolution.
Design and methods: We screened samples from 369 patients with newly diagnosed chronic lymphocytic leukemia from a population-based cohort using 250K single nucleotide polymorphism-arrays. Clonal evolution was evaluated in 59 follow-up samples obtained after 5-9 years.
Results: At diagnosis, copy-number aberrations were identified in 90% of patients; 70% carried known recurrent alterations, including del(13q) (55%), trisomy 12 (10.5%), del(11q) (10%), and del(17p) (4%). Additional recurrent aberrations were detected on chromosomes 2 (1.9%), 4 (1.4%), 8 (1.6%) and 14 (1.6%). Thirteen patients (3.5%) displayed recurrent copy-number neutral loss of heterozygosity on 13q, of whom 11 had concurrent homozygous del(13q). Genomic complexity and large 13q deletions correlated with inferior outcome, while the former was linked to poor-prognostic aberrations. In the follow-up study, clonal evolution developed in 8/24 (33%) patients with unmutated IGHV, and in 4/25 (16%) IGHV-mutated and treated patients. In contrast, untreated patients with mutated IGHV (n=10) did not acquire additional aberrations. The most common secondary event, del(13q), was detected in 6/12 (50%) of all patients with acquired alterations. Interestingly, aberrations on, for example, chromosome 6q, 8p, 9p and 10q developed exclusively in patients with unmutated IGHV.
Conclusions: Whole-genome screening revealed a high frequency of genomic aberrations in newly diagnosed chronic lymphocytic leukemia. Clonal evolution was associated with other markers of aggressive disease and commonly included the known recurrent aberrations.
Figures
Figure 1.
Genomic map of homozygous and heterozygous deletion 13q. (A) Chromosomal bands and location in Mbp, (B) genes and (C) a condensed view of the homozygous (light gray) and heterozygous (dark gray) deletions of the enlarged sub-region of deletion at 13q14.2–14.3. The single nucleotide polymorphisms defining the minimally deleted region (MDR) were rs706612 (49608579–49608580 bp) and rs1359612 (49612082-49612082 bp) for the centromeric breakpoint and rs1750567 (49635023–49635024 bp) and rs1798968 (49635869–49635870 bp) for the telomeric breakpoint.
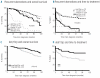
Figure 2.
Survival and time to treatment according to known recurrent aberrations and deletion 13q size (Kaplan-Meier plots). (A) Patients with del(17p) had the worst outcome, whereas del(11q) and trisomy 12 indicated similar intermediate survival. Patients with no recurrent aberration and patients with del(13q) showed similar good overall survival (P<0.0001). (B) Patients with poor-prognostic markers had a shorter time to treatment than patients with del(13q) and no recurrent aberration (_P_<0001). (C-D) Receiver operating characteristics (ROC) curve analysis and overall survival or time to treatment was applied in order to identify the optimal size cut-off for del(13q). Patients with larger del(13q) (>1.25 Mbp) had an inferior prognosis in terms of overall survival (C) (_P_=0.016), and time to treatment (D) (P<0.01).
Figure 3.
An increased genomic complexity predicts a worse survival and shorter time to treatment (Kaplan-Meier plots). Overall survival of patients was investigated according to the degree of genomic complexity, which showed (A) an inferior survival (P<0.0001) and (B) shorter time to treatment (P<0.0001) for patients carrying an increasing number of copy number aberrations larger than 5 Mbp.
Figure 4.
CLL patients with clonal evolution. (A-B) Examples of clonal evolution in two CLL patients for whom the first sample at diagnosis (upper panel) is compared to the follow-up sample in the lower panel of each figure. (A) SCAN 182 (IGHV unmutated and treated) had a normal copy-number of chromosome 10 at diagnosis, and developed a loss of the q-arm in the follow-up sample. (B) SCAN 75 (Binet stage A, IGHV unmutated) had no aberrations at diagnosis but showed a complex genome with deletions on 6q, 8p, 13q, 17p, 18p and a gain on 8q at follow-up after two lines of treatment with alkylating agents.
Similar articles
- High-density screening reveals a different spectrum of genomic aberrations in chronic lymphocytic leukemia patients with 'stereotyped' IGHV3-21 and IGHV4-34 B-cell receptors.
Marincevic M, Cahill N, Gunnarsson R, Isaksson A, Mansouri M, Göransson H, Rasmussen M, Jansson M, Ryan F, Karlsson K, Adami HO, Davi F, Jurlander J, Juliusson G, Stamatopoulos K, Rosenquist R. Marincevic M, et al. Haematologica. 2010 Sep;95(9):1519-25. doi: 10.3324/haematol.2009.021014. Epub 2010 Apr 26. Haematologica. 2010. PMID: 20421269 Free PMC article. - Importance of immunoglobulin heavy chain variable region mutational status in del(13q) chronic lymphocytic leukemia.
Gladstone DE, Swinnen L, Kasamon Y, Blackford A, Gocke CD, Griffin CA, Meade JB, Jones RJ. Gladstone DE, et al. Leuk Lymphoma. 2011 Oct;52(10):1873-81. doi: 10.3109/10428194.2011.585529. Epub 2011 Aug 18. Leuk Lymphoma. 2011. PMID: 21851216 Free PMC article. - Clinical, immunophenotypic, and molecular profiling of trisomy 12 in chronic lymphocytic leukemia and comparison with other karyotypic subgroups defined by cytogenetic analysis.
Athanasiadou A, Stamatopoulos K, Tsompanakou A, Gaitatzi M, Kalogiannidis P, Anagnostopoulos A, Fassas A, Tsezou A. Athanasiadou A, et al. Cancer Genet Cytogenet. 2006 Jul 15;168(2):109-19. doi: 10.1016/j.cancergencyto.2006.02.001. Cancer Genet Cytogenet. 2006. PMID: 16843100 - Exploring the genetic landscape in chronic lymphocytic leukemia using high-resolution technologies.
Gunnarsson R, Mansouri L, Rosenquist R. Gunnarsson R, et al. Leuk Lymphoma. 2013 Aug;54(8):1583-90. doi: 10.3109/10428194.2012.751530. Epub 2013 Feb 28. Leuk Lymphoma. 2013. PMID: 23167608 Review. - Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis.
Döhner H, Stilgenbauer S, Döhner K, Bentz M, Lichter P. Döhner H, et al. J Mol Med (Berl). 1999 Feb;77(2):266-81. doi: 10.1007/s001090050350. J Mol Med (Berl). 1999. PMID: 10023780 Review.
Cited by
- Uncovering the DNA methylome in chronic lymphocytic leukemia.
Cahill N, Rosenquist R. Cahill N, et al. Epigenetics. 2013 Feb;8(2):138-48. doi: 10.4161/epi.23439. Epub 2013 Jan 15. Epigenetics. 2013. PMID: 23321535 Free PMC article. Review. - Immunological and genetic kinetics from diagnosis to clinical progression in chronic lymphocytic leukemia.
Jiménez I, Tazón-Vega B, Abrisqueta P, Nieto JC, Bobillo S, Palacio-García C, Carabia J, Valdés-Mas R, Munuera M, Puigdefàbregas L, Parra G, Esteve-Codina A, Franco-Jarava C, Iacoboni G, Terol MJ, García-Marco JA, Crespo M, Bosch F. Jiménez I, et al. Biomark Res. 2021 May 20;9(1):37. doi: 10.1186/s40364-021-00290-z. Biomark Res. 2021. PMID: 34016160 Free PMC article. - Longitudinal copy number, whole exome and targeted deep sequencing of 'good risk' IGHV-mutated CLL patients with progressive disease.
Rose-Zerilli MJ, Gibson J, Wang J, Tapper W, Davis Z, Parker H, Larrayoz M, McCarthy H, Walewska R, Forster J, Gardiner A, Steele AJ, Chelala C, Ennis S, Collins A, Oakes CC, Oscier DG, Strefford JC. Rose-Zerilli MJ, et al. Leukemia. 2016 Jun;30(6):1301-10. doi: 10.1038/leu.2016.10. Epub 2016 Feb 5. Leukemia. 2016. PMID: 26847028 Free PMC article. - Genetic abnormalities in chronic lymphocytic leukemia: where we are and where we go.
Puiggros A, Blanco G, Espinet B. Puiggros A, et al. Biomed Res Int. 2014;2014:435983. doi: 10.1155/2014/435983. Epub 2014 May 22. Biomed Res Int. 2014. PMID: 24967369 Free PMC article. Review. - Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia.
Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN. Ouillette P, et al. Clin Cancer Res. 2013 Jun 1;19(11):2893-904. doi: 10.1158/1078-0432.CCR-13-0138. Epub 2013 Apr 25. Clin Cancer Res. 2013. PMID: 23620403 Free PMC article.
References
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. - PubMed
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. - PubMed
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–54. - PubMed
- Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720–4. - PubMed
- Juliusson G, Robert KH, Ost A, Friberg K, Biberfeld P, Nilsson B, et al. Prognostic information from cytogenetic analysis in chronic B-lymphocytic leukemia and leukemic immunocytoma. Blood. 1985;65(1):134–41. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases