Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death - PubMed (original) (raw)
Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death
Pieter J A de Koning et al. PLoS One. 2011.
Abstract
Granzyme-mediated cell death is the major pathway for cytotoxic lymphocytes to kill virus-infected and tumor cells. In humans, five different granzymes (i.e. GrA, GrB, GrH, GrK, and GrM) are known that all induce cell death. Expression of intracellular serine protease inhibitors (serpins) is one of the mechanisms by which tumor cells evade cytotoxic lymphocyte-mediated killing. Intracellular expression of SERPINB9 by tumor cells renders them resistant to GrB-induced apoptosis. In contrast to GrB, however, no physiological intracellular inhibitors are known for the other four human granzymes. In the present study, we show that SERPINB4 formed a typical serpin-protease SDS-stable complex with both recombinant and native human GrM. Mutation of the P2-P1-P1' triplet in the SERPINB4 reactive center loop completely abolished complex formation with GrM and N-terminal sequencing revealed that GrM cleaves SERPINB4 after P1-Leu. SERPINB4 inhibited GrM activity with a stoichiometry of inhibition of 1.6 and an apparent second order rate constant of 1.3×10(4) M(-1) s(-1). SERPINB4 abolished cleavage of the macromolecular GrM substrates α-tubulin and nucleophosmin. Overexpression of SERPINB4 in tumor cells inhibited recombinant GrM-induced as well as NK cell-mediated cell death and this inhibition depended on the reactive center loop of the serpin. As SERPINB4 is highly expressed by squamous cell carcinomas, our results may represent a novel mechanism by which these tumor cells evade cytotoxic lymphocyte-induced GrM-mediated cell death.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
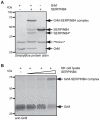
Figure 1. SERPINB4 forms a typical serpin-protease SDS-stable complex with recombinant and native human GrM.
(A) Purified recombinant GrM (1.8 µM) and SERPINB4 (1.8 µM) were incubated for 1 h at 37°C. All samples were treated with PNGase F, separated by SDS-PAGE, and analyzed by SimplyBlue staining. (B) Cell lysate of the NK cell line KHYG-1 (33 µg) was incubated with recombinant SERPINB4 (0, 50, 150, and 450 ng) for 2 h at 37°C. Subsequently, samples were immunoblotted for native GrM. GrM, SERPINB4, GrM-SERPINB4 complexes, and cleaved SERPINB4 (SERPINB4*) are indicated.
Figure 2. Mutation of the SERPINB4 RCL at the P2-P1-P1′ positions completely abolishes complex formation with human GrM.
(A) A RCL-mutant of SERPINB4 was employed in which the amino acids at the putative P2(Glu353)-P1(Leu354)-P1′(Ser355) positions were mutated into P2(Gln353)-P1(Gly354)-P1′(Ala355). Purified recombinant GrM (0.9 µM), GrM-SA (0.9 µM), SERPINB4 (0.9 µM), and SERPINB4 RCL-mutant (0.9 µM) were incubated for 1 h at 37°C. All samples were treated with PNGase F, separated by SDS-PAGE, and immunoblotted for GrM. Bound antibodies were visualized using DAB. (B) Cell lysates of 293T cells transfected with C-terminal GFP-conjugated SERPINB4 or an empty vector (mock) were incubated with recombinant GrM (0.5 µM) for 1 h at 37°C. Subsequently, samples were immunoblotted for GFP. SERPINB4-GFP represents the full length protein, whereas SERPINB4*-GFP depicts the C-terminal cleavage product. (C) Schematic representation of C-terminal GFP-conjugated SERPINB4, including the GrM cleavage site after Leu354 at the P1-position in the RCL.
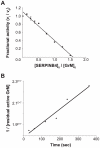
Figure 3. Kinetic analyses of GrM-inhibition by SERPINB4.
(A) Purified recombinant human GrM (2 µM) was incubated with different concentrations of recombinant SERPINB4 (0–3 µM) for 2 h at 37°C. Residual GrM activity was monitored by addition of a synthetic chromogenic leucine substrate (1 mM) and measuring A405 in time. The fractional activity (velocity of substrate hydrolysis by GrM in the presence of SERPINB4/velocity of substrate hydrolysis by GrM without SERPINB4) was plotted against the ratio of [SERPINB4]0/[GrM]0. Linear regression analysis was used to calculate the x-intercept as a value for the SI and determined to be 1.6±0.07 (mean ± SD of 4 independent experiments). (B) The apparent second order rate constant (kinh) of the inhibition of GrM by SERPINB4 was determined under second order conditions. Equimolar concentrations of recombinant human GrM and SERPINB4 were incubated. Aliquots were removed at different time points and the reaction was directly stopped by 20-fold dilution with buffer containing a synthetic chromogenic leucine substrate (1 mM). Residual GrM activity was determined by measuring the velocity of substrate hydrolysis in time at A405. These velocities were converted to residual active GrM concentrations using a GrM concentration standard curve. The apparent second order rate constant (kinh) was calculated from the slope of the plot of reciprocal residual active GrM over time and determined to be 1.3×104 M−1s−1 (graph indicates one representative example of three independent experiments with similar results).
Figure 4. SERPINB4 inhibits GrM-mediated cleavage of macromolecular substrates.
Indicated concentrations of recombinant GrM and SERPINB4 were incubated for 2 h at 37°C. Jurkat tumor cell lysate (2 µg) was added to the samples and incubated for another 4 h at 37°C to determine the residual GrM-activity towards macromolecular substrates. Finally, all samples were immunoblotted for α-tubulin, nucleophosmin, and β-actin.
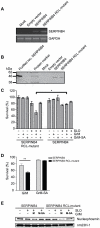
Figure 5. Overexpression of SERPINB4 in HeLa cells inhibits GrM-induced cell death.
(A) RT-PCR analysis of stably transfected HeLa cells for SERPINB4 and GAPDH mRNA expression (upper and lower panel, respectively). (B) Immunoblot analysis of SERPINB4 protein expression by HeLa cells stably transfected with pcDNA3 SERPINB4, pcDNA3 SERPINB4 RCL-mutant or pcDNA3 empty vector. (C) HeLa cells stably transfected with SERPINB4 or SERPINB4 RCL-mutant were treated with the indicated combinations of a sublytic dose of SLO (500 ng/ml), recombinant GrM (0.5 µM), and/or recombinant GrM-SA (0.5 µM) for 16 h at 37°C. Viable cells were quantified using the methylene blue assay. Data represent the percentages of viable cells as compared to HeLa cells that overexpressed SERPINB4 RCL-mutant and were treated with buffer only, which was set as 100%. Figure represents the mean ± SD of 4 independent experiments; * p<0.05. (D) HeLa cells stably transfected with SERPINB4 or SERPINB4 RCL-mutant were treated with the indicated combinations of a sublytic dose of SLO (500 ng/ml), recombinant GrM (1 µM), and/or recombinant GrM-SA (1 µM) for 20 h at 37°C. Cell viability was determined using flow cytometry, with AnnexinV and PI negative cells considered viable (mean ± S.D., n = 3, ** p<0.005). (E) HeLa cells stably transfected with SERPINB4 or SERPINB4 RCL-mutant were treated with the indicated combinations of a sublytic dose of SLO (500 ng/ml), recombinant GrM (1 µM), and/or recombinant GrM-SA (1 µM) for 4 h at 37°C. Total cell lysates were immunoblotted using antibodies against nucleophosmin and nm23H-1 (which served as a loading control).
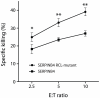
Figure 6. Overexpression of SERPINB4 in HeLa cells inhibits NK cell-mediated cell death.
HeLa cells stably transfected with SERPINB4 or SERPINB4 RCL-mutant were loaded with the fluorescent cell staining dye CFDA-SE and co-cultured with KHYG-1 NK cells in varying E∶T ratio's for 16 h at 37°C. Cells were stained with PI and analyzed by flow cytometry. HeLa cells were separated from KHYG-1 cells by gating for CFDA-SE positive cells. Depicted is the percentage of specific cytotoxicity (mean ± SD, n = 3, * p<0.05, ** p<0.005).
Similar articles
- A Novel Serpin Regulatory Mechanism: SerpinB9 IS REVERSIBLY INHIBITED BY VICINAL DISULFIDE BOND FORMATION IN THE REACTIVE CENTER LOOP.
Mangan MS, Bird CH, Kaiserman D, Matthews AY, Hitchen C, Steer DL, Thompson PE, Bird PI. Mangan MS, et al. J Biol Chem. 2016 Feb 12;291(7):3626-38. doi: 10.1074/jbc.M115.699298. Epub 2015 Dec 15. J Biol Chem. 2016. PMID: 26670609 Free PMC article. - NK cell protease granzyme M targets alpha-tubulin and disorganizes the microtubule network.
Bovenschen N, de Koning PJ, Quadir R, Broekhuizen R, Damen JM, Froelich CJ, Slijper M, Kummer JA. Bovenschen N, et al. J Immunol. 2008 Jun 15;180(12):8184-91. doi: 10.4049/jimmunol.180.12.8184. J Immunol. 2008. PMID: 18523284 - The cytotoxic protease granzyme M is expressed by lymphocytes of both the innate and adaptive immune system.
de Koning PJ, Tesselaar K, Bovenschen N, Colak S, Quadir R, Volman TJ, Kummer JA. de Koning PJ, et al. Mol Immunol. 2010 Jan;47(4):903-11. doi: 10.1016/j.molimm.2009.10.001. Epub 2009 Nov 5. Mol Immunol. 2010. PMID: 19896187 - Granzyme M: behind enemy lines.
de Poot SA, Bovenschen N. de Poot SA, et al. Cell Death Differ. 2014 Mar;21(3):359-68. doi: 10.1038/cdd.2013.189. Epub 2014 Jan 10. Cell Death Differ. 2014. PMID: 24413154 Free PMC article. Review. - Granzymes (lymphocyte serine proteases): characterization with natural and synthetic substrates and inhibitors.
Kam CM, Hudig D, Powers JC. Kam CM, et al. Biochim Biophys Acta. 2000 Mar 7;1477(1-2):307-23. doi: 10.1016/s0167-4838(99)00282-4. Biochim Biophys Acta. 2000. PMID: 10708866 Review.
Cited by
- Mechanisms of Apoptosis Resistance to NK Cell-Mediated Cytotoxicity in Cancer.
Sordo-Bahamonde C, Lorenzo-Herrero S, Payer ÁR, Gonzalez S, López-Soto A. Sordo-Bahamonde C, et al. Int J Mol Sci. 2020 May 25;21(10):3726. doi: 10.3390/ijms21103726. Int J Mol Sci. 2020. PMID: 32466293 Free PMC article. Review. - Integrative analysis of mutated genes and mutational processes reveals novel mutational biomarkers in colorectal cancer.
Dashti H, Dehzangi I, Bayati M, Breen J, Beheshti A, Lovell N, Rabiee HR, Alinejad-Rokny H. Dashti H, et al. BMC Bioinformatics. 2022 Apr 19;23(1):138. doi: 10.1186/s12859-022-04652-8. BMC Bioinformatics. 2022. PMID: 35439935 Free PMC article. - SERPINB3 (SCCA1) inhibits cathepsin L and lysoptosis, protecting cervical cancer cells from chemoradiation.
Wang S, Luke CJ, Pak SC, Shi V, Chen L, Moore J, Andress AP, Jayachandran K, Zhang J, Huang Y, Platik M, Apicelli AA, Schwarz JK, Grigsby PW, Silverman GA, Markovina S. Wang S, et al. Commun Biol. 2022 Jan 12;5(1):46. doi: 10.1038/s42003-021-02893-6. Commun Biol. 2022. PMID: 35022555 Free PMC article. - Squamous Cell Carcinoma Antigen 2 (SCCA2, SERPINB4): An Emerging Biomarker for Skin Inflammatory Diseases.
Izuhara K, Yamaguchi Y, Ohta S, Nunomura S, Nanri Y, Azuma Y, Nomura N, Noguchi Y, Aihara M. Izuhara K, et al. Int J Mol Sci. 2018 Apr 6;19(4):1102. doi: 10.3390/ijms19041102. Int J Mol Sci. 2018. PMID: 29642409 Free PMC article. Review. - Killer cell proteases can target viral immediate-early proteins to control human cytomegalovirus infection in a noncytotoxic manner.
Shan L, Li S, Meeldijk J, Blijenberg B, Hendriks A, van Boxtel KJWM, van den Berg SPH, Groves IJ, Potts M, Svrlanska A, Stamminger T, Wills MR, Bovenschen N. Shan L, et al. PLoS Pathog. 2020 Apr 13;16(4):e1008426. doi: 10.1371/journal.ppat.1008426. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32282833 Free PMC article.
References
- Barry M, Bleackley RC. Cytotoxic T lymphocytes: all roads lead to death. Nat Rev Immunol. 2002;2:401–409. - PubMed
- Smyth MJ, Kelly JM, Sutton VR, Davis JE, Browne KA, et al. Unlocking the secrets of cytotoxic granule proteins. J Leukoc Biol. 2001;70:18–29. - PubMed
- Cullen SP, Martin SJ. Mechanisms of granule-dependent killing. Cell Death Differ. 2008;15:251–262. - PubMed
- Bovenschen N, Kummer JA. Orphan granzymes find a home. Immunol Rev. 2010;235:117–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous