TIGRFAMs and Genome Properties in 2013 - PubMed (original) (raw)
. 2013 Jan;41(Database issue):D387-95.
doi: 10.1093/nar/gks1234. Epub 2012 Nov 28.
Affiliations
- PMID: 23197656
- PMCID: PMC3531188
- DOI: 10.1093/nar/gks1234
TIGRFAMs and Genome Properties in 2013
Daniel H Haft et al. Nucleic Acids Res. 2013 Jan.
Abstract
TIGRFAMs, available online at http://www.jcvi.org/tigrfams is a database of protein family definitions. Each entry features a seed alignment of trusted representative sequences, a hidden Markov model (HMM) built from that alignment, cutoff scores that let automated annotation pipelines decide which proteins are members, and annotations for transfer onto member proteins. Most TIGRFAMs models are designated equivalog, meaning they assign a specific name to proteins conserved in function from a common ancestral sequence. Models describing more functionally heterogeneous families are designated subfamily or domain, and assign less specific but more widely applicable annotations. The Genome Properties database, available at http://www.jcvi.org/genome-properties, specifies how computed evidence, including TIGRFAMs HMM results, should be used to judge whether an enzymatic pathway, a protein complex or another type of molecular subsystem is encoded in a genome. TIGRFAMs and Genome Properties content are developed in concert because subsystems reconstruction for large numbers of genomes guides selection of seed alignment sequences and cutoff values during protein family construction. Both databases specialize heavily in bacterial and archaeal subsystems. At present, 4284 models appear in TIGRFAMs, while 628 systems are described by Genome Properties. Content derives both from subsystem discovery work and from biocuration of the scientific literature.
Figures
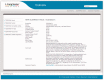
Figure 1.
Example of a TIGRFAMs web page. The TIGRFAMs homepage and all its web pages have five links located on the left navigational sidebar. These go to the TIGRFAMs Home page itself, a glossary of key terms used in TIGRFAMs, a complete listing of all models (4284 in release 13.0), the TIGRFAMs FTP Site, and a page with links to additional resources, including rapid searching of a protein sequence against HMM libraries. The page shown in the figure is summary page for a single TIGRFAMs entry, the Mb-OB3b-class methanobactin precursor family TIGR04071. Each summary page provides details about a single TIGRFAM, including cutoffs, HMM length, references and any assigned GO terms. The summary page also displays links to Genome Properties, if any, associated with that HMM. Users can view and download the HMM seed alignment through the HMM Seed Alignment link found on the left-hand side of the HMM Summary page. The TIGRFAMs complete listing page (not shown) displays accessions and functional names for all models, as well as length, isology type and EC number.
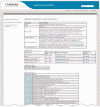
Figure 2.
Example of a Genome Properties Definition Page. The Genome Properties home page provides three links on the left navigation sidebar: Genome Properties List, Top Level Genome Properties and Search CMR Genome Properties. The Genome Properties List shows all current Genome Properties along with their property type and property name. Clicking on a Genome Property will display the Genome Property Definition page. The Definition Page for GenProp0962 (‘methanobactin biosynthesis, Mb-OB3b family’) is illustrated. The Genome Property Definition page displays the property’s name, description, parent/child/sibling properties, literature references and associated GO terms. The components of the property are listed with the step names, and whether or not the step is required (used to evaluate completeness). The evidence required to detect that a component has been found is described, usually the accession of a TIGRFAMs HMM with a working link to HMM Summary Page. Users can browse through the Genome Properties through the Top Level Genome Properties page linked on the home page. Users traverse through the properties by first selecting a top level property (e.g. Metabolism, Taxonomy). Associated parent, child and sibling properties are displayed for selection. The Search CMR Properties link directs users to the CMR genome property search.
Similar articles
- TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes.
Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. Selengut JD, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D260-4. doi: 10.1093/nar/gkl1043. Epub 2006 Dec 6. Nucleic Acids Res. 2007. PMID: 17151080 Free PMC article. - The TIGRFAMs database of protein families.
Haft DH, Selengut JD, White O. Haft DH, et al. Nucleic Acids Res. 2003 Jan 1;31(1):371-3. doi: 10.1093/nar/gkg128. Nucleic Acids Res. 2003. PMID: 12520025 Free PMC article. - TIGRFAMs: a protein family resource for the functional identification of proteins.
Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. Haft DH, et al. Nucleic Acids Res. 2001 Jan 1;29(1):41-3. doi: 10.1093/nar/29.1.41. Nucleic Acids Res. 2001. PMID: 11125044 Free PMC article. - The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST).
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. Overbeek R, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D206-14. doi: 10.1093/nar/gkt1226. Epub 2013 Nov 29. Nucleic Acids Res. 2014. PMID: 24293654 Free PMC article. - The SUPERFAMILY database in structural genomics.
Gough J. Gough J. Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1897-900. doi: 10.1107/s0907444902015160. Epub 2002 Oct 21. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12393919 Review.
Cited by
- The coral Oculina patagonica holobiont and its response to confinement, temperature, and Vibrio infections.
Martin-Cuadrado AB, Rubio-Portillo E, Rosselló F, Antón J. Martin-Cuadrado AB, et al. Microbiome. 2024 Oct 29;12(1):222. doi: 10.1186/s40168-024-01921-x. Microbiome. 2024. PMID: 39472959 Free PMC article. - Complete genomes of Asgard archaea reveal diverse integrated and mobile genetic elements.
Valentin-Alvarado LE, Shi LD, Appler KE, Crits-Christoph A, De Anda V, Adler BA, Cui ML, Ly L, Leão P, Roberts RJ, Sachdeva R, Baker BJ, Savage DF, Banfield JF. Valentin-Alvarado LE, et al. Genome Res. 2024 Oct 29;34(10):1595-1609. doi: 10.1101/gr.279480.124. Genome Res. 2024. PMID: 39406503 Free PMC article. - High-quality genome of a novel Thermosynechococcaceae species from Namibia and characterization of its protein expression patterns at elevated temperatures.
Arnold ND, Paper M, Fuchs T, Ahmad N, Jung P, Lakatos M, Rodewald K, Rieger B, Qoura F, Kandawa-Schulz M, Mehlmer N, Brück TB. Arnold ND, et al. Microbiologyopen. 2024 Oct;13(5):e70000. doi: 10.1002/mbo3.70000. Microbiologyopen. 2024. PMID: 39365014 Free PMC article. - Comprehensive analyses of a large human gut Bacteroidales culture collection reveal species- and strain-level diversity and evolution.
Zhang ZJ, Cole CG, Coyne MJ, Lin H, Dylla N, Smith RC, Pappas TE, Townson SA, Laliwala N, Waligurski E, Ramaswamy R, Woodson C, Burgo V, Little JC, Moran D, Rose A, McMillin M, McSpadden E, Sundararajan A, Sidebottom AM, Pamer EG, Comstock LE. Zhang ZJ, et al. Cell Host Microbe. 2024 Oct 9;32(10):1853-1867.e5. doi: 10.1016/j.chom.2024.08.016. Epub 2024 Sep 17. Cell Host Microbe. 2024. PMID: 39293438 - Interactive tools for functional annotation of bacterial genomes.
Price MN, Arkin AP. Price MN, et al. Database (Oxford). 2024 Sep 6;2024:baae089. doi: 10.1093/database/baae089. Database (Oxford). 2024. PMID: 39241109 Free PMC article.
References
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources