Inhibition of melanoma growth by small molecules that promote the mitochondrial localization of ATF2 - PubMed (original) (raw)
Inhibition of melanoma growth by small molecules that promote the mitochondrial localization of ATF2
Tal Varsano et al. Clin Cancer Res. 2013.
Abstract
Purpose: Effective therapy for malignant melanoma, the leading cause of death from skin cancer, remains an area of significant unmet need in oncology. The elevated expression of PKCε in advanced metastatic melanoma results in the increased phosphorylation of the transcription factor ATF2 on threonine 52, which causes its nuclear localization and confers its oncogenic activities. The nuclear-to-mitochondrial translocation of ATF2 following genotoxic stress promotes apoptosis, a function that is largely lost in melanoma cells, due to its confined nuclear localization. Therefore, promoting the nuclear export of ATF2, which sensitizes melanoma cells to apoptosis, represents a novel therapeutic modality.
Experimental design: We conducted a pilot high-throughput screen of 3,800 compounds to identify small molecules that promote melanoma cell death by inducing the cytoplasmic localization of ATF2. The imaging-based ATF2 translocation assay was conducted using UACC903 melanoma cells that stably express doxycycline-inducible GFP-ATF2.
Results: We identified two compounds (SBI-0089410 and SBI-0087702) that promoted the cytoplasmic localization of ATF2, reduced cell viability, inhibited colony formation, cell motility, and anchorage-free growth, and increased mitochondrial membrane permeability. SBI-0089410 inhibited the 12-O-tetradecanoylphorbol-l3-acetate (TPA)-induced membrane translocation of protein kinase C (PKC) isoforms, whereas both compounds decreased ATF2 phosphorylation by PKCε and ATF2 transcriptional activity. Overexpression of either constitutively active PKCε or phosphomimic mutant ATF2(T52E) attenuated the cellular effects of the compounds.
Conclusion: The imaging-based high-throughput screen provides a proof-of-concept for the identification of small molecules that block the oncogenic addiction to PKCε signaling by promoting ATF2 nuclear export, resulting in mitochondrial membrane leakage and melanoma cell death.
©2013 AACR
Conflict of interest statement
Conflict of interest: There are no conflicts to disclose.
Figures
Figure 1. The induction of GFP-ATF2 expression in the inducible GFP-ATF2Tet-Off UACC903 stable cell line and its application in an imaging-based HTP screen for inducers of ATF2 translocation
(A) GFP-ATF2 is expressed and readily detectable in GFP-ATF2Tet-Off UACC903 cells following removal of doxycycline from the medium. GFP-ATF2 expression was determined using direct fluorescence microscopy with an Olympus IX71 platform fitted with a 10× air objective lens (NA = 0.31). Scale bar = 20 μm. Insets are further magnified areas of each image. (B) Levels of induced GFP-ATF2 and endogenous ATF2 are comparable (1- to 5-fold) in the GFP-ATF2Tet-Off UACC903 stable cell line. Expression of GFP-ATF2 and endogenous ATF2 was compared in GFP-ATF2Tet-Off UACC903 cells and the parental cell line (negative control). GFP-ATF2Tet-Off UACC903 cells were plated and cultured in the presence or absence of 500 ng/ml doxycycline to inhibit or promote expression of GFP-ATF2, respectively. Cells were cultured for 12-72 h and lysed in RIPA buffer. Lysates were subjected to immunoblot analysis using rabbit anti-ATF2 and mouse anti-GFP antibodies. Actin and Ponceau staining were used for loading control. (C) Imaging-based assay read-out for ATF2 translocation. UACC903 cells stably expressing ATF2-GFP and that were stained with DAPI were imaged with the Opera QEHS system using DAPI and GFP wavelengths (top left and middle respectively, top right shows pseudo-colored overlay). (1) Nuclei detection from DAPI image via thresholding and object splitting (nuclei outlines in red). (2) Cell detection from contrast-stretched DAPI images via thresholding combined with nuclei location information (cell outlines in green). (3) The creation of cytoplasmic regions by subtraction of nuclei regions from the cell regions (cytoplasm outlines in yellow). (4) The overlay of nuclei (red) and cytoplasm (yellow) regions on ATF2 image. (5) Quantify nuclear metrics from nuclei regions of DAPI images and ATF2-based nuclei and cytoplasmic region metrics from ATF2-GFP images on a cell-by-cell level. (6) Create GFP-positive cell sub-population by applying a minimum intensity threshold for whole cell GFP intensity. (7) Calculate well-level assay plate read-outs from cell sub-population statistics of each well. Image analysis was performed using Acapella/Columbus HCS image analysis software.
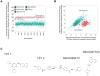
Figure 2. A pilot screen of a 3,800 compound ChemBridge library to identify compounds that promote the nuclear export of GFP-ATF2
(A) Summary of the results of the complete HTS screen showing the mean ratio of nuclear and cytoplasmic GFP fluorescence intensity for triplicate wells of each compound. (B) Nuclear and cytoplasmic intensities were also evaluated separately for each hit to remove compounds that interfere with the fluorescent protein directly or spectrally as indicated by a significant increase (Z-Score > 5) or decrease (Z-Score < 5) in both regions’ intensities simultaneously. Some autofluorescent compounds are outside of the axes range displayed here. The assay was designed away from the autofluroescent wavelength (Gö6850 is strongly autofluorescent in the red channel) by using green secondary antibodies and emission filters that would minimize the autofluorescence of the Go compound. The average Z’ for this screen was 0.47 and ranged between 0.4 and 0.56 in different plates. Cells were imaged from twelve 384-well plates (4,357 valid total wells 3,669 valid compound wells) using the Opera acquisition system and analyzed with Acapella software to identify GFP-positive cells and delineate nuclear and cytoplasmic regions. DMSO was used as the negative control (blue circles) and the PKC inhibitor Gö6850, which promotes nuclear export of ATF2, was used as the positive control (red circles). As expected, most of the compounds were inactive and exhibited ratios similar to the negative controls. (C) The structure of the 4 selected hits.
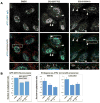
Figure 3. SBI-0089410 and SBI-0087702 promote the translocation of endogenous ATF2 to the cytoplasm/mitochondria
(A) Representative images of WM793 melanoma cells following treatment with DMSO, SBI-0089410, or SBI-0087702, showing increased localization of ATF2 to the cytoplasm and mitochondria (arrowheads). WM793 cells were cultured on glass coverslips and incubated with DMSO (0.2%, 24 h), SBI-0089410 (10 μM, 6 h), or SBI-0087702 (10 μM, 24 h). The cells were then fixed and stained with specific anti-ATF2 antibodies (green channel), anti-HSP60 antibodies to label mitochondria (red channel), and DAPI to label nuclei (blue channel). The cells were imaged using a FluoView1000 confocal microscope with an 100x oil immersion objective. Scale bar = 10 μm. Similar results were obtained with UACC903 cells (data not shown). (B) To measure ATF2 translocation, either the GFP fluorescence intensity of GFP-ATF2-expressing UACC903 (graph on left) or the ATF2 immunofluorescence intensity in WM793 and UACC903 cells (graphs on right) in the nucleus and cytoplasm were quantified using ImageJ software. Between 6 and 17 cells were analyzed for each condition. Quantification reveals a reduction of approximately 40-50% in the nuclear-to-cytoplasmic ATF2 ratio.
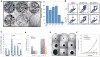
Figure 4. SBI-0089410 and SBI-0087702 inhibit colony formation, cell motility, anchorage-independent growth, and promote the apoptosis of melanoma cells in an ATF2-dependent manner
(A) Panels A-F in right image show representative wells containing colonies of 501Mel melanoma cells cultured in the presence of screen hits and controls as follows: (A) DMSO, (B) Gö6850, (C) hit 1, (D) hit 2, (E) SBI-0089410, and (F) SBI-0087702. 501Mel cells were plated at low density (500 cells/well in 6-well plates) and were grown in medium containing the indicated compounds. The number of colonies formed after 7 days in culture was determined by crystal violet staining. SBI-0089410 (410; E) and SBI-0087702 (702; F) were the most potent inhibitors of colony formation compared with Gö6850 (G). Similar results were obtained with the UACC903 cell line (data not shown). (B) Representative FACS profiles for pBabe empty vector (EV) or pBabe ATF2T52E (ATF2T52E)-stably transduced UACC903 cells that were treated with DMSO, 10 μM SBI-0089410 (410) or 10 μM SBI-0087702 (702) for 24 h. The cells were harvested, stained with Annexin V and propidium iodide (PI), and subjected to FACS analysis. N = 10,000 cells per replicate sample. (C) The histogram represents data from 3 independent experiments in (B) (ATF2T52E = 52E). (D) Quantification of the effect of hit compounds on the motility of WM1346 cell lines stably expressing phospho-mimic ATF2 mutant (ATF2 52E) or empty vector (EV). Expression of ATF2 enhanced cell motility by approximately 2-fold (right columns). Both hit compounds inhibited migration of EV cells but expression of ATF2 52E rescued the inhibitory effect. Cell motility was determined in a modified Boyden chamber assay using the Calcein-AM fluorecent staining as described in material and methods. The experiments were performed in triplicate wells and results show averages and standard deviations. (E) SBI-0089410 and SBI-0087702 inhibited the spheroid growth of SW1 melanoma cells. Representative images demonstrating the effect of hit compounds on SW1 spheroids growth. Spheroids were generated by the hanging drop method and subsequently transferred to separate wells and treated with the indicated compounds (10 μM) or vehicle (DMSO 0.08%) every other day for 8 days, in triplicates. Scale bar = 200 μm. (F) Quantification of spheroid growth showing average and standard deviation from 3 independent wells. Phase-contrast images of spheroids were obtained using an Olympus light microscope and spheroid diameter was measure using the SlideBook software. Similar results were obtained with UACC903 cells.
Figure 5. SBI-0089410 and SBI-0087702 promote mitochondrial leakage and reduced viability, which is block by expression of constitutively active PKCε
(A) SBI-0089410 (410) and SBI-0087702 (702) induce ATF2-dependent mitochondrial membrane leakage. WM793 cells were incubated DMSO alone, 10 μM hits 1 or 2 for 24 h, 10 μM SBI-0089410 for 6 and 24 h, 10 μM SBI-0087702 for 24 h, or with 50 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) for 45 min. Cell were then labeled with TMRE (250 μM) or NAO (10 nM) and analyzed by FACS. Samples were gated on whole cells by forward and side scatter and 10,000 gated cells were analyzed per sample. (B) Red FACS profiles represent TMRE uptake (reflective of mitochondrial membrane potential), whereas the green profiles represent NAO uptake (mitochondrial mass). Leftward peak shifts indicate decreased TMRE or NAO uptake, whereas rightward peak shifts indicate increased TMRE or NaO uptake. The dashed line indicates the median uptake values for the DMSO-treated samples. (C) WM793 cells were transiently transfected with control empty vector (EV) or ATF2T52E (pEF-HA-ATF2T52E) for 48 h. Cells were then treated with DMSO alone, 10 μM SBI-0089410 (410) for 6 h, 10 μM SBI-0087702 (702) for 24 h, or 50 μM CCCP for 45 min. The cells were labeled and analyzed as described for (A). Inset in (C): Western blot analysis showing HA-ATF2 expression. Histograms for (A) and (C) show the mean TMRE/NAO ratio values ± S.D. of 3 independent experiments. (D) Transient expression of a constitutively active form of PKCε (PKCε-CA) confers resistance to SBI-0087702-induced cell death. 501Mel melanoma cells were transiently transfected with either control GFP or PKCε-CA and incubated with the indicated compounds (10 μM final concentration) for 3 days before cell viability was measured using the CellTiter-Blue fluorescence assay (Promega). SBI-0087702-induced cell death was prevented by the expression of PKCε-CA.
Figure 6. SBI-0089410 and SBI-0087702 inhibit melanoma cell growth, ATF2 transcriptional activity, and affect the expression of ATF2 target genes in a PKCε-dependent manner
(A) NIH3T3 cells were incubated with DMSO, 10 μM SBI-0089410 (410) for 6 h, 10 μM SBI-0087702 (702) for 24 h, or with 10 μM of the PKCε inhibitor Gö6850. Anisomycin was then added for 30 min to prevent de novo protein synthesis and to activate cellular stress responses. Cells were lysed in RIPA buffer and analyzed by western blotting with the indicated antibodies. Protein bands were quantified using ImageJ software (numbers below). (B) SBI-0089410 (410) transiently inhibited ATF2 and p38 phosphorylation in UACC903 melanoma cells. Cells were pretreated with SBI-0089410 for 3-9 h, before stimulation with anisomycin. SBI-0089410 inhibited ATF2 phosphorylation at Thr52 and p38 phosphorylation with maximum activity after 6 h incubation. (C) SBI-0089410 inhibited the TPA-induced membrane translocation of PKC. WM793 cells were incubated in the presence of SBI-008410 or vehicle (DMSO) for 6 h, then stimulated with 100 nM TPA for 3 min, washed in cold PBS, lysed in lysis buffer, and the membrane and cytosol fractions were isolated according to the protocol detailed in the Material and Methods section. The different fractions were analyzed by Western blot using the antibodies indicated. (D) WM793 cells were treated with compounds and TPA as in (C) and subsequently fixed with 4% formaldehyde prior to processing for immunofluorescence using anti-PKCε primary and FITC-conjugated goat anti-rabbit-IgG secondary antibodies. Imaging was conducted using an Olympus IX-71 fluorescent microscope. Arrowheads point to PKCε staining at the plasma membrane and the insets show enlarged region near the plasma membrane. Scale-bar = 10 μm. (E) Luciferase assay evaluation of the effects of SBI-0089410 and SBI-0087702 on ATF2 transcriptional activity. UACC903 cells were stably transfected with a secreted 3X Jun2-(Gaussia) Luciferase construct. The cells were then transiently transfected with pCMV-Cypridina luciferase (normalization control), followed by treatment with DMSO, 10 μM SBI-0089410, 10 μM SBI-0087702, or with 10 μM PKCε translocation peptide inhibitor (PKCε-i). At the indicated time-points (0, 1, 6, 24 h = T0, T1, T6, T24), Gaussia and Cypridina luciferase activities were measured, and Gaussia luciferase activity was normalized to Cypridina luciferase activity. The graph represents the mean Jun2-luciferase activities ± S.D. relative to DMSO of 3 replicate experiments. (F) IPA analysis of gene expression changes observed upon treatment with SBI-0087702 identifies main clusters that are up- or downregulated. The lists of top 20 genes for each cluster are provided in Supplementary Tables 1 and 2. The original data were deposited into GEO (GSE43135).
Similar articles
- PKCε promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria.
Lau E, Kluger H, Varsano T, Lee K, Scheffler I, Rimm DL, Ideker T, Ronai ZA. Lau E, et al. Cell. 2012 Feb 3;148(3):543-55. doi: 10.1016/j.cell.2012.01.016. Cell. 2012. PMID: 22304920 Free PMC article. - ATF2 - at the crossroad of nuclear and cytosolic functions.
Lau E, Ronai ZA. Lau E, et al. J Cell Sci. 2012 Jun 15;125(Pt 12):2815-24. doi: 10.1242/jcs.095000. Epub 2012 Jun 8. J Cell Sci. 2012. PMID: 22685333 Free PMC article. - Novel small molecule XPO1/CRM1 inhibitors induce nuclear accumulation of TP53, phosphorylated MAPK and apoptosis in human melanoma cells.
Yang J, Bill MA, Young GS, La Perle K, Landesman Y, Shacham S, Kauffman M, Senapedis W, Kashyap T, Saint-Martin JR, Kendra K, Lesinski GB. Yang J, et al. PLoS One. 2014 Jul 24;9(7):e102983. doi: 10.1371/journal.pone.0102983. eCollection 2014. PLoS One. 2014. PMID: 25057921 Free PMC article. - ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities.
Bhoumik A, Ronai Z. Bhoumik A, et al. Cell Cycle. 2008 Aug;7(15):2341-5. doi: 10.4161/cc.6388. Epub 2008 Jun 17. Cell Cycle. 2008. PMID: 18677098 Review. - ATF2 on the double - activating transcription factor and DNA damage response protein.
Bhoumik A, Lopez-Bergami P, Ronai Z. Bhoumik A, et al. Pigment Cell Res. 2007 Dec;20(6):498-506. doi: 10.1111/j.1600-0749.2007.00414.x. Pigment Cell Res. 2007. PMID: 17935492 Free PMC article. Review.
Cited by
- Invasion-Block and S-MARVEL: A high-content screening and image analysis platform identifies ATM kinase as a modulator of melanoma invasion and metastasis.
Guo D, Jurek R, Beaumont KA, Sharp DS, Tan SY, Mariana A, Failes TW, Grootveld AK, Bhattacharyya ND, Phan TG, Arndt GM, Jain R, Weninger W, Tikoo S. Guo D, et al. Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2303978120. doi: 10.1073/pnas.2303978120. Epub 2023 Nov 14. Proc Natl Acad Sci U S A. 2023. PMID: 37963252 Free PMC article. - Pathways and therapeutic targets in melanoma.
Shtivelman E, Davies MQ, Hwu P, Yang J, Lotem M, Oren M, Flaherty KT, Fisher DE. Shtivelman E, et al. Oncotarget. 2014 Apr 15;5(7):1701-52. doi: 10.18632/oncotarget.1892. Oncotarget. 2014. PMID: 24743024 Free PMC article. Review. - Protein Kinase C (PKC) Isozymes as Diagnostic and Prognostic Biomarkers and Therapeutic Targets for Cancer.
Kawano T, Inokuchi J, Eto M, Murata M, Kang JH. Kawano T, et al. Cancers (Basel). 2022 Nov 3;14(21):5425. doi: 10.3390/cancers14215425. Cancers (Basel). 2022. PMID: 36358843 Free PMC article. Review. - ATF2, a paradigm of the multifaceted regulation of transcription factors in biology and disease.
Watson G, Ronai ZA, Lau E. Watson G, et al. Pharmacol Res. 2017 May;119:347-357. doi: 10.1016/j.phrs.2017.02.004. Epub 2017 Feb 15. Pharmacol Res. 2017. PMID: 28212892 Free PMC article. Review. - The Multifunctional Protein Kinase C-ε in Cancer Development and Progression.
Jain K, Basu A. Jain K, et al. Cancers (Basel). 2014 Apr 10;6(2):860-78. doi: 10.3390/cancers6020860. Cancers (Basel). 2014. PMID: 24727247 Free PMC article.
References
- Ko JM, Fisher DE. A new era: melanoma genetics and therapeutics. The Journal of pathology. 2011;223:241–50. - PubMed
- Shepherd C, Puzanov I, Sosman JA. B-RAF inhibitors: an evolving role in the therapy of malignant melanoma. Current oncology reports. 2010;12:146–52. - PubMed
- Smalley KS. Understanding melanoma signaling networks as the basis for molecular targeted therapy. The Journal of investigative dermatology. 2010;130:28–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous