miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs - PubMed (original) (raw)
Marijn T M van Jaarsveld, Antoinette Hollestelle, Wendy J C Prager-van der Smissen, Anouk A J Heine, Antonius W M Boersma, Jingjing Liu, Jean Helmijr, Bahar Ozturk, Marcel Smid, Erik A Wiemer, John A Foekens, John W M Martens
- PMID: 23601657
- PMCID: PMC3672661
- DOI: 10.1186/bcr3415
miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs
Muhammad Riaz et al. Breast Cancer Res. 2013.
Abstract
Introduction: Breast cancer is a genetically and phenotypically complex disease. To understand the role of miRNAs in this molecular complexity, we performed miRNA expression analysis in a cohort of molecularly well-characterized human breast cancer cell lines to identify miRNAs associated with the most common molecular subtypes and the most frequent genetic aberrations.
Methods: Using a microarray carrying LNA™ modified oligonucleotide capture probes), expression levels of 725 human miRNAs were measured in 51 breast cancer cell lines. Differential miRNA expression was explored by unsupervised cluster analysis and was then associated with the molecular subtypes and genetic aberrations commonly present in breast cancer.
Results: Unsupervised cluster analysis using the most variably expressed miRNAs divided the 51 breast cancer cell lines into a major and a minor cluster predominantly mirroring the luminal and basal intrinsic subdivision of breast cancer cell lines. One hundred and thirteen miRNAs were differentially expressed between these two main clusters. Forty miRNAs were differentially expressed between basal-like and normal-like/claudin-low cell lines. Within the luminal-group, 39 miRNAs were associated with ERBB2 overexpression and 24 with E-cadherin gene mutations, which are frequent in this subtype of breast cancer cell lines. In contrast, 31 miRNAs were associated with E-cadherin promoter hypermethylation, which, contrary to E-cadherin mutation, is exclusively observed in breast cancer cell lines that are not of luminal origin. Thirty miRNAs were associated with p16INK4 status while only a few miRNAs were associated with BRCA1, PIK3CA/PTEN and TP53 mutation status. Twelve miRNAs were associated with DNA copy number variation of the respective locus.
Conclusion: Luminal-basal and epithelial-mesenchymal associated miRNAs determine the subdivision of miRNA transcriptome of breast cancer cell lines. Specific sets of miRNAs were associated with ERBB2 overexpression, p16INK4a or E-cadherin mutation or E-cadherin methylation status, which implies that these miRNAs may contribute to the driver role of these genetic aberrations. Additionally, miRNAs, which are located in a genomic region showing recurrent genetic aberrations, may themselves play a driver role in breast carcinogenesis or contribute to a driver gene in their vicinity. In short, our study provides detailed molecular miRNA portraits of breast cancer cell lines, which can be exploited for functional studies of clinically important miRNAs.
Figures
Figure 1
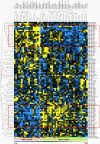
Molecular subtyping of 51 human breast cancer cell lines. Pearson correlation plot based on global mRNA expression of the top 10% variable genes. Breast cancer cell lines are depicted according to their overall similarity in gene expression. Yellow and blue, high and low overall similarity of samples in mRNA expression, respectively. Two main groups of 27 and 23 cell lines were apparent. Color codes for breast cancer subtypes based on intrinsic gene set: green, luminal-type cell lines; brown, luminal ERBB2-positive cell lines; black, normal-like/claudin-low cell lines; orange, basal-like cell lines; blue, estrogen receptor (ER)-negative/ERBB2-positive cell lines; pink, other subtype cell lines.
Figure 2
Global miRNA expression analysis of 51 human breast cancer cell lines. Unsupervised clustering across the breast cancer cell lines using the 87 most variably expressed miRNAs. Yellow and blue, high expression and low expression of the particular miRNA in the particular sample, respectively. Cell lines in the dendrogram of the hierarchical clustering based on miRNA expression are coded according to their intrinsic subtypes and depicted at the bottom of the figure (for color codes see Figure 1 caption). Red boxes enlist the groups of miRNAs that show the most robust cluster-specific differential expression in the cell lines. miRNA hsa-miR-21 in the red box of minor cluster failed to reach our criteria of fold change ≥ 1.5.
Figure 3
Molecular subtype-specific differential expression of miRNAs. Differentially expressed miRNAs (A) between luminal-type and luminal ERBB2-positive breast cancer cell lines within the mRNA-derived luminal-group (see Figure 1), and (B) between basal-like and normal-like/claudin-low breast cancer cell lines within the mRNA-derived estrogen receptor-negative/basal-group. Yellow and blue, high and low overall similarity of samples in miRNA expression, respectively.
Figure 4
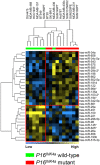
Differential expression of miRNAs with respect to E-cadherin status. Differentially expressed miRNAs (A) between E-cadherin mutant and wild-type breast cancer cell lines in the luminal-group, and (B) between E-cadherin promoter hypermethylated and wild-type breast cancer cell lines in the estrogen receptor-negative/basal group. Cell lines HCC1806, SK-BR-7, SUM102PT, SUM149PT, and SUM229PE, which show partial promoter hypermethylation, were not included in the analysis since we were not sure to which extent partial promoter methylation will affect E-cadherin expression levels in these cell lines. Yellow and blue, high and low overall similarity of samples in miRNA expression, respectively.
Figure 5
Differentially expressed miRNAs between p16INK4a mutant and wild-type cell lines in the estrogen receptor-negative/basal-group. The analysis was restricted to only the estrogen receptor (ER)-negative/basal-group of cell lines since the majority of p16INK4a mutant cell lines were ER-negative. Yellow and blue, high and low overall similarity of samples in miRNA expression, respectively.
Figure 6
Association of miRNA expression with genomic copy number variation in breast cancer cell lines. The top four most significant miRNAs are represented (see Table S16 in Additional file 1 for a complete list). The Kruskal-Wallis test was used to reveal significant associations of miRNAs with genome copy number (CN) variation. y axis, expression levels of miRNA; x axis, number of samples with CN loss/gain or neutral.
Similar articles
- Molecular features of the basal-like breast cancer subtype based on BRCA1 mutation status.
Prat A, Cruz C, Hoadley KA, Díez O, Perou CM, Balmaña J. Prat A, et al. Breast Cancer Res Treat. 2014 Aug;147(1):185-91. doi: 10.1007/s10549-014-3056-x. Epub 2014 Jul 22. Breast Cancer Res Treat. 2014. PMID: 25048467 Free PMC article. - Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics.
Jönsson G, Staaf J, Vallon-Christersson J, Ringnér M, Holm K, Hegardt C, Gunnarsson H, Fagerholm R, Strand C, Agnarsson BA, Kilpivaara O, Luts L, Heikkilä P, Aittomäki K, Blomqvist C, Loman N, Malmström P, Olsson H, Johannsson OT, Arason A, Nevanlinna H, Barkardottir RB, Borg A. Jönsson G, et al. Breast Cancer Res. 2010;12(3):R42. doi: 10.1186/bcr2596. Epub 2010 Jun 24. Breast Cancer Res. 2010. PMID: 20576095 Free PMC article. - Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer.
McDermott AM, Miller N, Wall D, Martyn LM, Ball G, Sweeney KJ, Kerin MJ. McDermott AM, et al. PLoS One. 2014 Jan 31;9(1):e87032. doi: 10.1371/journal.pone.0087032. eCollection 2014. PLoS One. 2014. PMID: 24498016 Free PMC article. - Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes.
Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J. Kurozumi S, et al. J Hum Genet. 2017 Jan;62(1):15-24. doi: 10.1038/jhg.2016.89. Epub 2016 Jul 21. J Hum Genet. 2017. PMID: 27439682 Review. - MicroRNAs in breast cancer pathogenesis.
Götte M. Götte M. Minerva Ginecol. 2010 Dec;62(6):559-71. Minerva Ginecol. 2010. PMID: 21079577 Review.
Cited by
- Gynaecological cancers and their cell lines.
Skok K, Gradišnik L, Maver U, Kozar N, Sobočan M, Takač I, Arko D, Kavalar R. Skok K, et al. J Cell Mol Med. 2021 Apr;25(8):3680-3698. doi: 10.1111/jcmm.16397. Epub 2021 Mar 2. J Cell Mol Med. 2021. PMID: 33650759 Free PMC article. Review. - The Targeted Degradation of BRAF V600E Reveals the Mechanisms of Resistance to BRAF-Targeted Treatments in Colorectal Cancer Cells.
Chapdelaine AG, Ku GC, Sun G, Ayrapetov MK. Chapdelaine AG, et al. Cancers (Basel). 2023 Dec 12;15(24):5805. doi: 10.3390/cancers15245805. Cancers (Basel). 2023. PMID: 38136350 Free PMC article. - Dumbbell-PCR: a method to quantify specific small RNA variants with a single nucleotide resolution at terminal sequences.
Honda S, Kirino Y. Honda S, et al. Nucleic Acids Res. 2015 Jul 13;43(12):e77. doi: 10.1093/nar/gkv218. Epub 2015 Mar 16. Nucleic Acids Res. 2015. PMID: 25779041 Free PMC article. - Development of a Novel Humanized Monoclonal Antibody to Secreted Frizzled-Related Protein-2 That Inhibits Triple-Negative Breast Cancer and Angiosarcoma Growth In Vivo.
Garcia D, Nasarre P, Bonilla IV, Hilliard E, Peterson YK, Spruill L, Broome AM, Hill EG, Yustein JT, Mehrotra S, Klauber-DeMore N. Garcia D, et al. Ann Surg Oncol. 2019 Dec;26(13):4782-4790. doi: 10.1245/s10434-019-07800-2. Epub 2019 Sep 12. Ann Surg Oncol. 2019. PMID: 31515721 Free PMC article. - Molecular Characteristics and Therapeutic Vulnerabilities of Claudin-low Breast Cancers Derived from Cell Line Models.
Voutsadakis IA. Voutsadakis IA. Cancer Genomics Proteomics. 2023 Nov-Dec;20(6):539-555. doi: 10.21873/cgp.20404. Cancer Genomics Proteomics. 2023. PMID: 37889067 Free PMC article.
References
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. - PubMed
- Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, Mäkelä R, Naume B, Perälä M, Kallioniemi O, Kristensen VN, Yakhini Z, Børresen-Dale AL. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous