CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect rRNA synthesis - PubMed (original) (raw)
. 2013 Jul-Aug;4(4):315-25.
doi: 10.4161/nucl.25342. Epub 2013 Jun 12.
Affiliations
- PMID: 23782956
- PMCID: PMC3810339
- DOI: 10.4161/nucl.25342
CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect rRNA synthesis
Baoyan Bai et al. Nucleus. 2013 Jul-Aug.
Abstract
CRM1 is an export factor that together with its adaptor NMD3 transports numerous cargo molecules from the nucleus to cytoplasm through the nuclear pore. Previous studies have suggested that CRM1 and NMD3 are detected in the nucleolus. However, their localization with subnucleolar domains or participation in the activities of the nucleolus are unclear. We demonstrate here biochemically and using imaging analyses that CRM1 and NMD3 co-localize with nucleolar marker proteins in the nucleolus. In particular, their nucleolar localization is markedly increased by inhibition of RNA polymerase I (Pol I) transcription by actinomycin D or by silencing Pol I catalytic subunit, RPA194. We show that CRM1 nucleolar localization is dependent on its activity and the expression of NMD3, whereas NMD3 nucleolar localization is independent of CRM1. This suggests that NMD3 provides nucleolar tethering of CRM1. While inhibition of CRM1 by leptomycin B inhibited processing of 28S ribosomal (r) RNA, depletion of NMD3 did not, suggesting that their effects on 28S rRNA processing are distinct. Markedly, depletion of NMD3 and inhibition of CRM1 reduced the rate of pre-47S rRNA synthesis. However, their inactivation did not lead to nucleolar disintegration, a hallmark of Pol I transcription stress, suggesting that they do not directly regulate transcription. These results indicate that CRM1 and NMD3 have complex functions in pathways that couple rRNA synthetic and processing engines and that the rRNA synthesis rate may be adjusted according to proficiency in rRNA processing and export.
Keywords: biogenesis; export; nucleolus; rRNA synthesis; transcription.
Figures
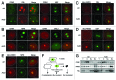
Figure 1. CRM1 and Myc-NMD3 accumulate in the nucleolus. (A) HeLa cells were treated with ActD (50 ng/ml) for 3 h as indicated. The cells were co-stained for FBL and CRM1 or for UBF and CRM1 as shown. (B) Cells expressing NPM-ECGFP were treated with ActD followed by staining for CRM1, or HeLa cells were treated with ActD followed by co-staining for GNL3 and CRM1. (C and D) HeLa cells transfected with Myc-NMD3 expression vector were treated as in (A) and co-stained for Myc-NMD3 and FBL (C) or Myc-NMD3 and UBF (D). (E) HeLa cells stably expressing NPM-ECGFP were transfected with Myc-NMD3, treated as in (A) and stained for Myc-NMD3. Scale bars, 10 µm. (F) Protocol of cellular fractionation to purify cytoplasmic, C; nuclear, Nu and nucleolar, No fractions. Note that the nuclear fraction contains also the nucleoli. T denotes total cellular extract. (G) Cells were treated with Act D (50 ng/ml) for 3 h followed by fractionation according to the scheme in (F), followed by western blot analysis for CRM1 and NMD3. Forty micrograms of protein from each fraction was loaded. FBL was used as a nucleolar marker.
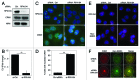
Figure 2. Depletion of RPA194 increases CRM1 and NMD3 nucleolar localization. (A) HeLa cells were transfected with control or RPA194 targeting siRNAs and incubated for 48 h, followed by western analysis for RPA194 and CRM1, and NPM was used as a loading control. (B) rRNA synthesis was determined using EU incorporation. Control and RPA194 siRNA transfected cells were incubated for 1 h with EU. EU incorporation was determined by quantitative image analysis. Data represents means ± SD, n = 2 experiments. ** p < 0.01. (C) RPA194 depleted cells or control siRNA transfected cells were immunostained for RPA194 and CRM1. Scale bar, 10 µm. (D) Quantification of number of cells with CRM1 nucleolar accumulation from (C). Data represents means ± SD, n = 2 experiments. ** p < 0.01. (E) Cells were transfected with siRNAs as indicated, followed by transfection with Myc-NMD3 expression vector and incubation for 24 h. Cells were fixed and stained for RPA194 and Myc-NMD3. Scale bar, 10 µm. (F) Cells treated as in (E) were co-stained for CRM1 and Myc-NMD3. CRM1, red; Myc-NMD, green; Merge, yellow. Scale bar, 10 µm.
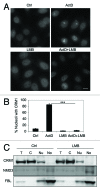
Figure 3. CRM1 nucleolar localization depends on its activity. (A) HeLa cells were treated with ActD (40 nM) or LMB (20 ng/ml) or their combination for 3 h, fixed and stained for CRM1. Scale bar, 10 µm. (B) Quantification of cells with CRM1 nucleolar accumulation in A. Data represents means ± SD, n = 2 experiments. *** p < 0.001. (C) Cells were treated with LMB (20 ng/ml) for 12 h followed by cellular fractionation and isolation of proteins as in Figure 1F. Protein loading was adjusted and analyzed by western blotting for CRM1, NMD3 and FBL.
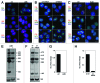
Figure 4. Myc-NMD3 nucleolar localization is independent of CRM1. (A and B) Cells were depleted of RPA194 using RNAi, transfected with (A) Myc-NMD3 or (B) Myc-NMD3-NES expression vectors and treated with LMB as shown for 4 h. Cells were fixed and stained for Myc-NMD3 using Myc-epitope tag antibody. Scale bar, 10 µm. (C) Quantification of nucleolar expression of Myc-tagged NMD3 and Myc-NMD3-NES. The percentage of cells with nucleolar expression is shown. Data represents means ± SD, n = 2 experiments.
Figure 5. CRM1 nucleolar accumulation is dependent on NMD3. (A) Cells were transfected with control or NMD3 targeting siRNAs and incubated for 48 h, followed by Western analysis for NMD3 and CRM1. (B) Quantitative PCR for the expression of NMD3 transcript. qPCR was normalized to GAPDH. (C) Cells were silenced for NMD3 as in A and treated with ActD (50 ng/ml) for 3 h and stained for CRM1. Scale bars, 10 µm. (D) Quantification of cells with CRM1 nucleolar accumulation in (C). Data represents means ± SD, n = 2 experiments. **, p < 0.001.
Figure 6. Alterations in nucleolar rRNAs following ActD and LMB treatments and NMD3 silencing. (A) Schematic presentation of rRNA processing. (B) RNA gel analysis of total RNA of cellular fractions treated with ActD (50 ng/ml) for 3 h. RNA was extracted from the samples in Figure 1G. EtBr staining. The migration of rRNAs is indicated to the right. (C) Cells were treated with LMB (20 ng/ml) for 12 h followed by cellular fractionation and isolation of RNA. The RNA gel was stained with EtBr (top panel). Subsequently, the RNA agarose gel was hybridized to digoxigenin-labeled 28S LNA probe (Northern, bottom panel). RNA was extracted from the samples in Figure 3C. (D) Cells were treated with ActD followed by in situ hybridization using probes detecting 5′ETS and 28S rRNAs. Quantification of the image analysis (right panel). Scale bar, 10 µm. (E) Cells were treated with LMB followed by in situ hybridization using probes detecting 5′ETS and 28S rRNA. Quantification of the image analysis (right panel). Scale bar, 10 µm. (F) RNA gel analysis of total RNA of cellular fractions from cells transfected with control or NMD3 siRNAs. EtBr staining.
Figure 7. Inhibition of CRM1 and NMD3 affects rRNA synthesis. (A, B and C) Immunofluorescence analysis of the indicated nucleolar proteins following treatment of cells with LMB for 14 h (A), NMD3 silencing (B) or treatment of the cells with ActD for 3 h (C). Scale bars, 10 µm. (D) Cells were treated with LMB for 4 h and labeled for the last two hours with 3H-uridine. RNA was isolated and separated by agarose gel electrophoresis. Autoradiogram is shown. (E) Cells were transfected with or without NMD3 silencing siRNAs and metabolic labeling of rRNA was conducted as in (D). Position of rRNAs are indicated. Total 18S rRNA as stained by EtBr is shown at the bottom. (F and G) Cells were treated with LMB for 13 h as in (D) or NMD3 was silenced as in (E) and cells were incubated for the last hour with EU. EU incorporation was detected, cells were imaged and nucleolar EU signals were quantified of over 200 cells in each assay. n = 2. Error bars, SD. Data represents means ± SD, n = 2 experiments.
Similar articles
- Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway.
Thomas F, Kutay U. Thomas F, et al. J Cell Sci. 2003 Jun 15;116(Pt 12):2409-19. doi: 10.1242/jcs.00464. Epub 2003 Apr 30. J Cell Sci. 2003. PMID: 12724356 - Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex.
West M, Hedges JB, Lo KY, Johnson AW. West M, et al. J Biol Chem. 2007 May 11;282(19):14028-37. doi: 10.1074/jbc.M700256200. Epub 2007 Mar 8. J Biol Chem. 2007. PMID: 17347149 - Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates.
Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. Trotta CR, et al. EMBO J. 2003 Jun 2;22(11):2841-51. doi: 10.1093/emboj/cdg249. EMBO J. 2003. PMID: 12773398 Free PMC article. - CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes.
Verheggen C, Bertrand E. Verheggen C, et al. Nucleus. 2012 Mar 1;3(2):132-7. doi: 10.4161/nucl.19266. Epub 2012 Mar 1. Nucleus. 2012. PMID: 22555597 Free PMC article. Review. - Joining the interface: a site for Nmd3 association on 60S ribosome subunits.
Oeffinger M. Oeffinger M. J Cell Biol. 2010 Jun 28;189(7):1071-3. doi: 10.1083/jcb.201006033. J Cell Biol. 2010. PMID: 20584913 Free PMC article. Review.
Cited by
- Uncovering the assembly pathway of human ribosomes and its emerging links to disease.
Bohnsack KE, Bohnsack MT. Bohnsack KE, et al. EMBO J. 2019 Jul 1;38(13):e100278. doi: 10.15252/embj.2018100278. Epub 2019 May 14. EMBO J. 2019. PMID: 31268599 Free PMC article. Review. - mRNA export in the apicomplexan parasite Toxoplasma gondii: emerging divergent components of a crucial pathway.
Ávila AR, Cabezas-Cruz A, Gissot M. Ávila AR, et al. Parasit Vectors. 2018 Jan 25;11(1):62. doi: 10.1186/s13071-018-2648-4. Parasit Vectors. 2018. PMID: 29370868 Free PMC article. Review. - Role of Rsp5 ubiquitin ligase in biogenesis of rRNA, mRNA and tRNA in yeast.
Domanska A, Kaminska J. Domanska A, et al. RNA Biol. 2015;12(12):1265-74. doi: 10.1080/15476286.2015.1094604. RNA Biol. 2015. PMID: 26403176 Free PMC article. Review. - Disruption of nucleocytoplasmic trafficking as a cellular senescence driver.
Park JH, Ryu SJ, Kim BJ, Cho HJ, Park CH, Choi HJC, Jang EJ, Yang EJ, Hwang JA, Woo SH, Lee JH, Park JH, Choi KM, Kwon YY, Lee CK, Park JT, Cho SC, Lee YI, Lee SB, Han JA, Cho KA, Kim MS, Hwang D, Lee YS, Park SC. Park JH, et al. Exp Mol Med. 2021 Jun;53(6):1092-1108. doi: 10.1038/s12276-021-00643-6. Epub 2021 Jun 29. Exp Mol Med. 2021. PMID: 34188179 Free PMC article. - XPO1 inhibitor KPT-330 synergizes with Bcl-xL inhibitor to induce cancer cell apoptosis by perturbing rRNA processing and Mcl-1 protein synthesis.
Zhu ZC, Liu JW, Yang C, Zhao M, Xiong ZQ. Zhu ZC, et al. Cell Death Dis. 2019 May 21;10(6):395. doi: 10.1038/s41419-019-1627-9. Cell Death Dis. 2019. PMID: 31113936 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous