Human gut microbiota changes reveal the progression of glucose intolerance - PubMed (original) (raw)
Human gut microbiota changes reveal the progression of glucose intolerance
Xiuying Zhang et al. PLoS One. 2013.
Abstract
To explore the relationship of gut microbiota with the development of type 2 diabetes (T2DM), we analyzed 121 subjects who were divided into 3 groups based on their glucose intolerance status: normal glucose tolerance (NGT; n = 44), prediabetes (Pre-DM; n = 64), or newly diagnosed T2DM (n = 13). Gut microbiota characterizations were determined with 16S rDNA-based high-throughput sequencing. T2DM-related dysbiosis was observed, including the separation of microbial communities and a change of alpha diversity between the different glucose intolerance statuses. To assess the correlation between metabolic parameters and microbiota diversity, clinical characteristics were also measured and a significant association between metabolic parameters (FPG, CRP) and gut microbiota was found. In addition, a total of 28 operational taxonomic units (OTUs) were found to be related to T2DM status by the Kruskal-Wallis H test, most of which were enriched in the T2DM group. Butyrate-producing bacteria (e.g. Akkermansia muciniphila ATCCBAA-835, and Faecalibacterium prausnitzii L2-6) had a higher abundance in the NGT group than in the pre-DM group. At genus level, the abundance of Bacteroides in the T2DM group was only half that of the NGT and Pre-DM groups. Previously reported T2DM-related markers were also compared with the data in this study, and some inconsistencies were noted. We found that Verrucomicrobiae may be a potential marker of T2DM as it had a significantly lower abundance in both the pre-DM and T2DM groups. In conclusion, this research provides further evidence of the structural modulation of gut microbiota in the pathogenesis of diabetes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
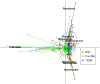
Figure 1. Principal component analysis (PCA) analysis of the similarity of microbiota (OTUs) between the NGT, Pre-DM, and T2DM groups.
Data for NGT (n = 44), Pre-DM (n = 64) and T2DM (n = 13) subjects were plotted on the first two principal components of the OTU profiles. The first 2 components (contributing 19.5% of variance) were plotted. Lines connect individuals belonging to the same group and colored circles cover individuals near the center of gravity for each cluster (<1.5_σ_). The top 7 OTUs (labeled with their annotations) for the main contributors to these groups were determined and plotted by their loadings for these 2 components. PCA was performed with R package ‘ade4’. OTU = operational taxonomic unit.
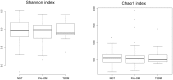
Figure 2. Alpha diversity of the 3 groups.
The Shannon index (left panel) and Chao1 index (right panel) were computed for all 121 subjects. The box depicts the interquartile range (IQR) between the first and third quartiles (25th and 75th percentiles, respectively) and the line inside the box denotes the median.
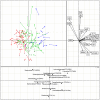
Figure 3. Co-inertia analysis (CIA) of the relationship between microbiota at the OTU level, clinical parameters, and disease group.
The upper left panel shows the CIA of the clinical parameter principal component analysis (PCA) and the microbiota PCA; arrows indicate where samples in the clinical parameter dataset are relative to the microbiota dataset. Red lines represent the NGT group, green lines the Pre-DM group, and blue lines the T2DM group. The upper right panel shows clinical parameter loading data; the lower panel displays the associated microbiota at the OTU level (labeled with their annotations). Only OTUs present in at least 1% of the samples were used in the analysis. CIA was performed with R package ‘ade4’. OTU = operational taxonomic unit.
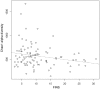
Figure 4. Correlations between the fasting insulin (FINS) concentration (mIU/L) and microbiota diversity.
The dashed line was derived from a simple regression model (_P_-value for the model was 0.034; R = −0.22).
Similar articles
- Metagenomics and Faecal Metabolomics Integrative Analysis towards the Impaired Glucose Regulation and Type 2 Diabetes in Uyghur-Related Omics.
Nuli R, Azhati J, Cai J, Kadeer A, Zhang B, Mohemaiti P. Nuli R, et al. J Diabetes Res. 2019 Nov 18;2019:2893041. doi: 10.1155/2019/2893041. eCollection 2019. J Diabetes Res. 2019. PMID: 31828159 Free PMC article. - Gut Microbiota Composition in Prediabetes and Newly Diagnosed Type 2 Diabetes: A Systematic Review of Observational Studies.
Letchumanan G, Abdullah N, Marlini M, Baharom N, Lawley B, Omar MR, Mohideen FBS, Addnan FH, Nur Fariha MM, Ismail Z, Pathmanathan SG. Letchumanan G, et al. Front Cell Infect Microbiol. 2022 Aug 15;12:943427. doi: 10.3389/fcimb.2022.943427. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36046745 Free PMC article. Review. - Aberrant intestinal microbiota in individuals with prediabetes.
Allin KH, Tremaroli V, Caesar R, Jensen BAH, Damgaard MTF, Bahl MI, Licht TR, Hansen TH, Nielsen T, Dantoft TM, Linneberg A, Jørgensen T, Vestergaard H, Kristiansen K, Franks PW; IMI-DIRECT consortium; Hansen T, Bäckhed F, Pedersen O. Allin KH, et al. Diabetologia. 2018 Apr;61(4):810-820. doi: 10.1007/s00125-018-4550-1. Epub 2018 Jan 29. Diabetologia. 2018. PMID: 29379988 Free PMC article. - Characterization of the gut microbiota in leptin deficient obese mice - Correlation to inflammatory and diabetic parameters.
Ellekilde M, Krych L, Hansen CH, Hufeldt MR, Dahl K, Hansen LH, Sørensen SJ, Vogensen FK, Nielsen DS, Hansen AK. Ellekilde M, et al. Res Vet Sci. 2014 Apr;96(2):241-50. doi: 10.1016/j.rvsc.2014.01.007. Epub 2014 Feb 3. Res Vet Sci. 2014. PMID: 24556473 - Vitamin D and prebiotics may benefit the intestinal microbacteria and improve glucose homeostasis in prediabetes and type 2 diabetes.
Barengolts E. Barengolts E. Endocr Pract. 2013 May-Jun;19(3):497-510. doi: 10.4158/EP12263.RA. Endocr Pract. 2013. PMID: 23823585 Review.
Cited by
- Gut microbiota composition and type 2 diabetes: Are these subjects linked Together?
Razavi S, Amirmozafari N, Zahedi Bialvaei A, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Sedighi M. Razavi S, et al. Heliyon. 2024 Oct 16;10(20):e39464. doi: 10.1016/j.heliyon.2024.e39464. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39469674 Free PMC article. - Effects of Artemisia asiatica ex on Akkermansia muciniphila dominance for modulation of Alzheimer's disease in mice.
Lee M, Ahn KS, Kim M. Lee M, et al. PLoS One. 2024 Oct 28;19(10):e0312670. doi: 10.1371/journal.pone.0312670. eCollection 2024. PLoS One. 2024. PMID: 39466764 Free PMC article. - Self-Initiated Dietary Adjustments Alter Microbiota Abundances: Implications for Perceived Health.
Willems A, Sura-de Jong M, Klaassens E, van den Bogert B, van Beek A, van Dijk G. Willems A, et al. Nutrients. 2024 Oct 18;16(20):3544. doi: 10.3390/nu16203544. Nutrients. 2024. PMID: 39458538 Free PMC article. - Harnessing Prebiotics to Improve Type 2 Diabetes Outcomes.
Iatcu OC, Hamamah S, Covasa M. Iatcu OC, et al. Nutrients. 2024 Oct 11;16(20):3447. doi: 10.3390/nu16203447. Nutrients. 2024. PMID: 39458444 Free PMC article. Review. - Causal relationship between gut microbiota and insulin-like growth factor 1: a bidirectional two-sample Mendelian randomization study.
Zheng X, Qian Y, Wang L. Zheng X, et al. Front Cell Infect Microbiol. 2024 Sep 24;14:1406132. doi: 10.3389/fcimb.2024.1406132. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39386166 Free PMC article.
References
- Yang W, Lu J, Weng J, Jia W, Ji L, et al. (2010) China National Diabetes and Metabolic Disorders Study Group. Prevalence of diabetes among men and women in China. N Engl J Med 362: 1090–1101. - PubMed
- Diamond Project Group (2006) Incidence and trends of childhood type 1 diabetes worldwide 1990–1999. Diabet Med 23: 857–866. - PubMed
- Coutinho M, Gerstein HC, Wang Y, Yusuf S (1999) The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes Care 22: 233–240. - PubMed
- Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, et al. (2008) Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 359: 2220–2232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
Financial support from the National High Technology Research and Development Program (2006AA02A409). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous