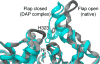Nonredox nickel enzymes - PubMed (original) (raw)
Review
. 2014 Apr 23;114(8):4206-28.
doi: 10.1021/cr4004488. Epub 2013 Dec 26.
Affiliations
- PMID: 24369791
- PMCID: PMC5675112
- DOI: 10.1021/cr4004488
Review
Nonredox nickel enzymes
Michael J Maroney et al. Chem Rev. 2014.
Abstract
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures
Figure 1
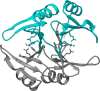
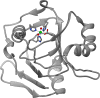
(A) Ribbon scheme of the functional oligomer (αβγ)3 of B. pasteurii urease. (B) Ribbon scheme of the functional oligomer (αβγ)3 of K. aerogenes urease. (C) Ribbon scheme of the functional oligomer [(αβ)3]4 of H. pylori urease seen through the ternary axis (left panel) and rotated by 90° along the horizontal axis (right panel). (D) Ribbon scheme of the functional oligomer [(α)3]2 of C. ensiformis urease seen through the ternary axis (left panel) and rotated by 90° along the horizontal axis (right panel).
Figure 2
Ribbon scheme of the active site flap of B. pasteurii urease, highlighting the open and closed conformations observed in the native and the DAP-inhibited structures, respectively.
Figure 3
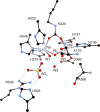
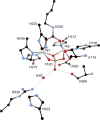
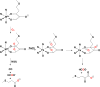
CrystalMaker drawing of the crystallographic structural models for the active site obtained for B. pasteurii urease (PDB code 2UBP) in the native state. The nickel ions are represented in gray, while CPK coloring is used for all other atoms. Hydrogen bonds are shown as thin blue lines. The BPU residue-numbering scheme (all residues belonging to the α subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 4
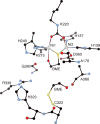
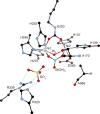
CrystalMaker drawing of the crystallographic structural model for the active site obtained for B. pasteurii urease complexed with _β_-mercaptoethanol (BME) (PDB code 1UBP). The nickel ions are represented in gray, while CPK coloring is used for all other atoms. Hydrogen bonds are shown as thin blue lines. The BPU residue-numbering scheme (all residues belonging to the α subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 5
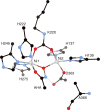
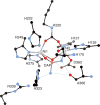
CrystalMaker drawing of the crystallographic structural model for the active site obtained for urease from B. pasteurii complexed with acetohydroxamic acid (AHA) (PDB code 4UBP). The nickel ions are represented in gray, while CPK coloring is used for all other atoms. Hydrogen bonds are shown as thin blue lines. The BPU residue-numbering scheme (all residues belonging to the α subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 6
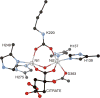
CrystalMaker drawing of the crystallographic structural model for the active site obtained for B. pasteurii urease complexed with phosphate (PHO) (PDB code 1IE7). The nickel ions are represented in gray and phosphorus is in orange, while CPK coloring is used for all other atoms. WAT = solvent molecule. Hydrogen bonds are shown as thin blue lines. The BPU residue-numbering scheme (all residues belonging to the α subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 7
CrystalMaker drawing of the crystallographic structural model for the active site obtained for B. pasteurii urease complexed with boric acid B(OH)3 (PDB code 1S3T). The nickel ions are represented in gray and boron is in green, while CPK coloring is used for all other atoms. WB = nickel-bridging hydroxide. Hydrogen bonds are shown as thin blue lines. The BPU residue-numbering scheme (all residues belonging to the α subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 8
CrystalMaker drawing of the crystallographic structural model for the active site obtained for B. pasteurii urease complexed with diaminophosphate (DAP) (PDB code 3UBP). The nickel ions are represented in gray and phosphorus is in orange, while CPK coloring is used for all other atoms. Hydrogen bonds are shown as thin blue lines. The BPU residue-numbering scheme (all residues belonging to the α subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 9
CrystalMaker drawing of the crystallograhic structural model for the active site obtained for B. pasteurii urease complexed with citrate (PDB code 4AC7). The nickel ions are represented in gray, while CPK coloring is used for all other atoms. The BPU residue-numbering scheme (all residues belonging to the alpha subunit) is used. The residue indicated with the letter “K” is the carbamylated lysine.
Figure 10
Structure-based urease catalytic mechanism of the enzymatic hydrolysis of urea. The BPU residue-numbering scheme is used.
Figure 11
Sequence alignments of selected class I and class II glyoxalase I enzymes created using Clustal W2. Amino acids are colored by property (hydrophobic (red), acidic (blue), basic (purple), other (green)). Metal binding residues are highlighted in yellow. Residues marked with an asterisk (∗) are invariant; those marked by other symbols represent low (:) and moderate (.) variability. The N-terminal extension and additional loops found in class I enzymes are highlighted in blue. The S. cerevisiae sequence was truncated after 226 of 326 residues.
Figure 12
Ribbon diagram of the crystal structure of E. coli Glo I, (PDB code 1F9Z) showing the two subunits of the homo dimer in cyan and gray and the location of the two Ni sites (green spheres) at subunit interfaces.
Figure 13
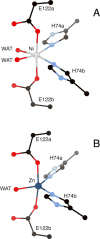
Comparison of the metal site structure of the Ni(II) complex (panel A, PDB code 1F9Z) and the Zn(II) complex (panel B, 1FA5) of E. coli Glo I showing the change in coordination number and geometry for the two metals. The nickel and zinc ions are represented in gray and dark blue, respectively, while CPK coloring is used for all other atoms. WAT = solvent molecules. Protein residues are distinguished by letters indicating the two different subunits of the enzyme.
Figure 14
Putative reaction mechanism for the isomerization catalyzed by Glo I that involves coordination of the substrate.
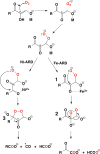
Figure 15
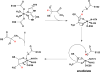
Simplified diagram of the methionine salvage pathway, illustrating key products, intermediates, and catalysis by Ni-ARD vs Fe-ARD.
Figure 16
Sequence alignments of selected ARD enzymes created using Clustal W2. The sequences are numbered from Met0, since this residue is cleaved in the mature enzyme. Amino acids are colored by property (hydrophobic (red), acidic (blue), basic (purple), other (green)). Metal binding residues are highlighted in yellow. Residues marked with an asterisk (∗) are invariant; those marked by other symbols represent low (:) and moderate (.) variability.
Figure 17
Ribbon diagram of the NMR structure of K. oxytoca Ni-ARD (PDB code 1ZRR) showing the cupin fold and the location of the metal ion (green sphere) with the ligand environment shown as sticks.
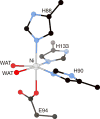
Figure 18
Metal site structure of K. oxytoca Ni-ARD (PDB code 1ZRR), showing the His3Glu coordination of the metal site and the two _cis_-aqua ligands in the positions used in binding substrate. The nickel ion is represented in gray, while CPK coloring is used for all other atoms. WAT = solvent molecules.
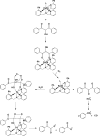
Figure 19
Proposed reaction mechanism illustrating the chelate hypothesis to explain the regioselectivity of the reactions catalyzed by Ni-ARD vs Fe-ARD. The results of incorporation of 18O and 14C labeling studies are indicated by the red O atoms and the asterisks (∗). (Adapted with permission from ref . Copyright 2007 John Wiley & Sons, Ltd.)
Figure 20
Model chemistry illustrating the role of substrate hydration in determining the regioselectivity of the reactions catalyzed by Ni-ARD vs Fe-ARD.
Figure 21
Mechanisms for Ni-ARD vs Fe-ARD catalysis from computational modeling indicate that the electronic structure of the metal ions leads to additional intermediates in the Fe-ARD reaction pathway.
Scheme 1
Scheme 2
Similar articles
- Nickel-dependent metalloenzymes.
Boer JL, Mulrooney SB, Hausinger RP. Boer JL, et al. Arch Biochem Biophys. 2014 Feb 15;544:142-52. doi: 10.1016/j.abb.2013.09.002. Epub 2013 Sep 10. Arch Biochem Biophys. 2014. PMID: 24036122 Free PMC article. Review. - Structure, function, and biosynthesis of nickel-dependent enzymes.
Alfano M, Cavazza C. Alfano M, et al. Protein Sci. 2020 May;29(5):1071-1089. doi: 10.1002/pro.3836. Epub 2020 Feb 18. Protein Sci. 2020. PMID: 32022353 Free PMC article. Review. - Distinct classes of glyoxalase I: metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes.
Sukdeo N, Clugston SL, Daub E, Honek JF. Sukdeo N, et al. Biochem J. 2004 Nov 15;384(Pt 1):111-7. doi: 10.1042/BJ20041006. Biochem J. 2004. PMID: 15270717 Free PMC article. - An evolutionary approach to the design of glutathione-linked enzymes.
Mannervik B, Cameron AD, Fernandez E, Gustafsson A, Hansson LO, Jemth P, Jiang F, Jones TA, Larsson AK, Nilsson LO, Olin B, Pettersson PL, Ridderström M, Stenberg G, Widersten M. Mannervik B, et al. Chem Biol Interact. 1998 Apr 24;111-112:15-21. doi: 10.1016/s0009-2797(97)00147-6. Chem Biol Interact. 1998. PMID: 9679539 Review. - Bacterial glyoxalase I enzymes: structural and biochemical investigations.
Honek JF. Honek JF. Biochem Soc Trans. 2014 Apr;42(2):479-84. doi: 10.1042/BST20130285. Biochem Soc Trans. 2014. PMID: 24646264
Cited by
- MTM1 displays a new function in the regulation of nickel resistance in Saccharomyces cerevisiae.
Xu N, Xu Y, Smith N, Chen H, Guo Z, Lee J, Wu X. Xu N, et al. Metallomics. 2022 Oct 8;14(10):mfac074. doi: 10.1093/mtomcs/mfac074. Metallomics. 2022. PMID: 36138538 Free PMC article. - Schiff bases and their metal complexes as urease inhibitors - A brief review.
de Fátima Â, Pereira CP, Olímpio CRSDG, de Freitas Oliveira BG, Franco LL, da Silva PHC. de Fátima Â, et al. J Adv Res. 2018 Mar 26;13:113-126. doi: 10.1016/j.jare.2018.03.007. eCollection 2018 Sep. J Adv Res. 2018. PMID: 30094086 Free PMC article. Review. - Soyuretox, an Intrinsically Disordered Polypeptide Derived from Soybean (Glycine Max) Ubiquitous Urease with Potential Use as a Biopesticide.
Kappaun K, Martinelli AHS, Broll V, Zambelli B, Lopes FC, Ligabue-Braun R, Fruttero LL, Moyetta NR, Bonan CD, Carlini CR, Ciurli S. Kappaun K, et al. Int J Mol Sci. 2019 Oct 30;20(21):5401. doi: 10.3390/ijms20215401. Int J Mol Sci. 2019. PMID: 31671552 Free PMC article. - Inhibition of Urease by Hydroquinones: A Structural and Kinetic Study.
Mazzei L, Cianci M, Ciurli S. Mazzei L, et al. Chemistry. 2022 Nov 16;28(64):e202201770. doi: 10.1002/chem.202201770. Epub 2022 Sep 22. Chemistry. 2022. PMID: 35994380 Free PMC article. - Inhibition of Urease by Disulfiram, an FDA-Approved Thiol Reagent Used in Humans.
Díaz-Sánchez ÁG, Alvarez-Parrilla E, Martínez-Martínez A, Aguirre-Reyes L, Orozpe-Olvera JA, Ramos-Soto MA, Núñez-Gastélum JA, Alvarado-Tenorio B, de la Rosa LA. Díaz-Sánchez ÁG, et al. Molecules. 2016 Nov 26;21(12):1628. doi: 10.3390/molecules21121628. Molecules. 2016. PMID: 27898047 Free PMC article.
References
- Sigel A, Sigel H, Sigel RKO, editors. Nickel and Its Surprising Impact on Nature. John Wiley & Sons, Ltd.; Chichester, England: 2007.
- Zambelli B, Ciurli S. Nickel and human health. In: Sigel A, Sigel H, Sigel RKO, editors. Interrelations between Essential Metal Ions and Human Diseases. Vol. 13 Springer Science and Business Media B.V.; Dordrecht, Germany: 2014.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources