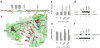The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes - PubMed (original) (raw)
Li Dong 2, Xiang Chen 3, Gang Wu 3, Matthew Parker 3, Lei Wei 3, Jing Ma 4, Michael N Edmonson 3, Erin K Hedlund 3, Michael C Rusch 3, Sheila A Shurtleff 4, Heather L Mulder 5, Kristy Boggs 5, Bhavin Vadordaria 5, Jinjun Cheng 4, Donald Yergeau 5, Guangchun Song 4, Jared Becksfort 3, Gordon Lemmon 3, Catherine Weber 4, Zhongling Cai 4, Jinjun Dang 4, Michael Walsh 6, Amanda L Gedman 4, Zachary Faber 4, John Easton 5, Tanja Gruber 7, Richard W Kriwacki 8, Janet F Partridge 9, Li Ding 10, Richard K Wilson 10, Elaine R Mardis 10, Charles G Mullighan 4, Richard J Gilbertson 11, Suzanne J Baker 11, Gerard Zambetti 9, David W Ellison 4, Jinghui Zhang 3, James R Downing 4
Affiliations
- PMID: 24710217
- PMCID: PMC4119022
- DOI: 10.1038/ncomms4630
The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes
Robert Huether et al. Nat Commun. 2014.
Abstract
Studies of paediatric cancers have shown a high frequency of mutation across epigenetic regulators. Here we sequence 633 genes, encoding the majority of known epigenetic regulatory proteins, in over 1,000 paediatric tumours to define the landscape of somatic mutations in epigenetic regulators in paediatric cancer. Our results demonstrate a marked variation in the frequency of gene mutations across 21 different paediatric cancer subtypes, with the highest frequency of mutations detected in high-grade gliomas, T-lineage acute lymphoblastic leukaemia and medulloblastoma, and a paucity of mutations in low-grade glioma and retinoblastoma. The most frequently mutated genes are H3F3A, PHF6, ATRX, KDM6A, SMARCA4, ASXL2, CREBBP, EZH2, MLL2, USP7, ASXL1, NSD2, SETD2, SMC1A and ZMYM3. We identify novel loss-of-function mutations in the ubiquitin-specific processing protease 7 (USP7) in paediatric leukaemia, which result in decreased deubiquitination activity. Collectively, our results help to define the landscape of mutations in epigenetic regulatory genes in paediatric cancer and yield a valuable new database for investigating the role of epigenetic dysregulations in cancer.
Conflict of interest statement
Competing Financial interests
The authors declare no competing financial interests.
Figures
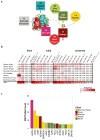
Figure 1. The landscape of somatic mutations in epigenetic regulators in 21 pediatric cancer subtypes
(a) Eight classes of epigenetic genes were interrogated across the cohort (Histone Writer, Bind Histone Writer, Histone Eraser, Bind Histone Eraser, Histone, Histone Reader, Chromatin Modifier, and DNA modifier), with the numbers of genes within each class indicated. (b) Fraction of tumors in each cancer subtype with at least one mutation in each class of epigenetic genes. Only sequence mutations (i.e. SNVs and indels) with a mutant allele fraction >0.3 (i.e. present in the dominant clone) were included in the analysis (c) Top 15 most frequently mutated genes colored coded by class.
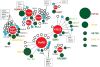
Figure 2. Epigenetic complexes affected by recurrently mutated proteins in pediatric cancer
A subset (35%) of the recurrently mutated epigenetic regulatory proteins (green circles) function within one or more of eight key epigenetic protein complexes (red nodes). Individual somatic mutation were also detected in additional components of these complexes (blue circles), whereas other components were never found to be mutated within our patient cohort (white circles). The size of each green circle is proportional to the number of mutated samples. The distance between the circles and the central complex node indicates whether the protein is a core (short) or transient (long) component of the complex. Recurrently mutated proteins that do not belong to one of these core complexes are presented on the right as unattached circles. The color of each protein name conforms to the color scheme for epigenetic regulatory classes presented in Figure 1.
Figure 3. Novel ALL-specific mutations of USP7
(a) Location of the identified USP7 somatic mutations relative to the TRAF (tumor necrosis factor [TNF] receptor-associated factor), catalytic, and HUBL1-5 (USP7/HAUSP ubiquitin-like domain) domains (colored red, green, black, orange, teal, purple and blue respectively). Mutations C300R, D483fs, and Q821R occurred at mutant allele frequencies (MAF) below 30%, whereas all other mutations occurred with MAF >30% and thus represent the dominant malignant clone. (b) Location of the missense somatic mutations (C300R, D305G, and A381T: magenta space filled) within the USP7 catalytic domain (green cartoon) – Ubiquitin (peach cartoon) interface. Specific residues and interactions between USP7 and Ubiquitin are shown as sticks and black dots and further described in Supplementary Fig. 3–5. (c and d) 293T cells were transfected with USP7 WT or mutant constructs as indicated. Protein extracts were prepared at 72 hours post transfection and subjected to western blot analysis using antibodies to the indicated proteins. Bars represent mean of protein band intensities of 3 replicates ± S.E.M. (e and f). The level of Histone H2B ubiquityl Lys120 (H2BK120ub1) and total H2B were detected at 72 hours by immunoblot using an antibody specific for mono-ubiquitinated and total H2B. Bars represent mean of protein band intensities of 3 replicates ± S.E.M. NT, untransfected control. The statistical significance of the changes observed between wild type and USP7 mutants were assessed by t-test with * equal to a p<0.05 and ** equal to p<0.01.
Similar articles
- Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia.
Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, Kung AL, Lo Nigro L, Ebert BL, Armstrong SA. Mar BG, et al. Nat Commun. 2014 Mar 24;5:3469. doi: 10.1038/ncomms4469. Nat Commun. 2014. PMID: 24662245 Free PMC article. - Invited Review: Emerging functions of histone H3 mutations in paediatric diffuse high-grade gliomas.
Kasper LH, Baker SJ. Kasper LH, et al. Neuropathol Appl Neurobiol. 2020 Feb;46(1):73-85. doi: 10.1111/nan.12591. Epub 2020 Jan 7. Neuropathol Appl Neurobiol. 2020. PMID: 31859390 Free PMC article. Review. - Epigenetic regulation in medulloblastoma.
Yi J, Wu J. Yi J, et al. Mol Cell Neurosci. 2018 Mar;87:65-76. doi: 10.1016/j.mcn.2017.09.003. Epub 2017 Dec 18. Mol Cell Neurosci. 2018. PMID: 29269116 Free PMC article. Review. - Tumour immune landscape of paediatric high-grade gliomas.
Ross JL, Velazquez Vega J, Plant A, MacDonald TJ, Becher OJ, Hambardzumyan D. Ross JL, et al. Brain. 2021 Oct 22;144(9):2594-2609. doi: 10.1093/brain/awab155. Brain. 2021. PMID: 33856022 Free PMC article. Review. - CREBBP mutations in relapsed acute lymphoblastic leukaemia.
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, Buetow KH, Pui CH, Baker SD, Brindle PK, Downing JR. Mullighan CG, et al. Nature. 2011 Mar 10;471(7337):235-9. doi: 10.1038/nature09727. Nature. 2011. PMID: 21390130 Free PMC article.
Cited by
- Large-Scale Identification of Clonal Hematopoiesis and Mutations Recurrent in Blood Cancers.
Feusier JE, Arunachalam S, Tashi T, Baker MJ, VanSant-Webb C, Ferdig A, Welm BE, Rodriguez-Flores JL, Ours C, Jorde LB, Prchal JT, Mason CC. Feusier JE, et al. Blood Cancer Discov. 2021 May;2(3):226-237. doi: 10.1158/2643-3230.BCD-20-0094. Epub 2021 Mar 3. Blood Cancer Discov. 2021. PMID: 34027416 Free PMC article. - Chromatin regulators in retinoblastoma: Biological roles and therapeutic applications.
Lee C, Kim JK. Lee C, et al. J Cell Physiol. 2021 Apr;236(4):2318-2332. doi: 10.1002/jcp.30022. Epub 2020 Aug 25. J Cell Physiol. 2021. PMID: 32840881 Free PMC article. Review. - Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations.
Roberti A, Dobay MP, Bisig B, Vallois D, Boéchat C, Lanitis E, Bouchindhomme B, Parrens MC, Bossard C, Quintanilla-Martinez L, Missiaglia E, Gaulard P, de Leval L. Roberti A, et al. Nat Commun. 2016 Sep 7;7:12602. doi: 10.1038/ncomms12602. Nat Commun. 2016. PMID: 27600764 Free PMC article. - Targeted Next-Generation Sequencing Identifies Actionable Targets in Estrogen Receptor Positive and Estrogen Receptor Negative Endometriod Endometrial Cancer.
Suhaimi SS, Ab Mutalib NS, Khor SS, Zain RRM, Syafruddin SE, Abu N, Mohd Dali AZH, Jamal R. Suhaimi SS, et al. Front Pharmacol. 2018 Jul 13;9:750. doi: 10.3389/fphar.2018.00750. eCollection 2018. Front Pharmacol. 2018. PMID: 30057548 Free PMC article. - Chromatin-Independent Interplay of NFATc1 and EZH2 in Pancreatic Cancer.
Patil S, Forster T, Reutlinger K, Kopp W, Versemann L, Spitalieri J, Gaedcke J, Ströbel P, Singh SK, Ellenrieder V, Neesse A, Hessmann E. Patil S, et al. Cells. 2021 Dec 8;10(12):3463. doi: 10.3390/cells10123463. Cells. 2021. PMID: 34943970 Free PMC article.
References
- Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous