Integrating canonical and metabolic signalling programmes in the regulation of T cell responses - PubMed (original) (raw)
Review
Integrating canonical and metabolic signalling programmes in the regulation of T cell responses
Kristen N Pollizzi et al. Nat Rev Immunol. 2014 Jul.
Abstract
Over the past decade, our understanding of T cell activation, differentiation and function has markedly expanded, providing a greater appreciation of the signals and pathways that regulate these processes. It has become clear that evolutionarily conserved pathways that regulate stress responses, metabolism, autophagy and survival have crucial and specific roles in regulating T cell responses. Recent studies suggest that the metabolic pathways involving MYC, hypoxia-inducible factor 1α (HIF1α), AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) are activated upon antigen recognition and that they are required for directing the consequences of T cell receptor engagement. The purpose of this Review is to provide an integrated view of the role of these metabolic pathways and of canonical T cell signalling pathways in regulating the outcome of T cell responses.
Figures
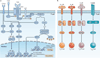
Figure 1. Canonical T cell signalling pathways: signal 1 and signal 2
a | Signal 1 (T cell receptor (TCR) engagement) in the setting of signal 2 (co-stimulation; depicted as CD28) leads to full T cell activation. This is facilitated by the activation of three canonical transcription factors — nuclear factor-κB (NF-κB), activator protein 1 (AP-1) and nuclear factor of activated T cells (NFAT),,,. This, in turn, leads to the expression of multiple cytokines, chemokines and cell surface receptors, all of which promote T cell activation and proliferation. Alternatively, TCR recognition alone (in the absence of co-stimulation) leads to an ‘off’ signal in the form of T cell anergy,. Under these conditions, NFAT is activated in the absence of full AP-1 activation, which leads to the expression of genes such as diacylglycerol kinase-α (DGKA) and the E3 ubiquitin-protein ligases CBLB and GRAIL (which encodes gene related to anergy in lymphocytes; also known as RNF128), which inhibit full T cell activation. b | Upon T cell activation, cytokines in the T cell microenvironment determine the outcome of antigen recognition with regard to effector T cell differentiation. As shown for CD4+ T cells, interleukin-12 (IL-12), IL-4 and IL-6 activate signal transducer and activator of transcription 4 (STAT4), STAT6 and STAT3, respectively. This leads to the expression of T-bet, GATA-binding protein 3 (GATA3) and retinoic acid receptor-related orphan receptor-γt (RORγt), which facilitates the generation of T helper 1 (TH1), TH2 and TH17 cells. Alternatively, transforming growth factor-β (TGFβ) signalling through SMAD2-SMAD4 promotes the expression of forkhead box P3 (FOXP3) and the generation of regulatory T (TReg) cells. DAG, diacylglycerol; IKK, inhibitor of NF-κB kinase; InsP3, inositol-1,4,5-trisphosphate; LAT, linker for activation of T cells; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; PI3K, phosphoinositide 3-kinase; PKCθ, protein kinase Cθ; PLCγ, phospholipase Cγ; PtdInsP2, phosphatidylinositol-4,5-bisphosphate; SLP76, SH2 domain-containing leukocyte protein of 76 kDa (also known as LCP2); ZAP70, ζ-chain-associated protein kinase of 70 kDa.
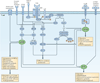
Figure 2. Integrating immunological and metabolic signalling programmes to promote effector T cell generation and function
The figure shows the coordinated integration of canonical T cell signalling (blue) and metabolic regulators (green) to promote the generation and function of effector T cells. In this perspective, hypoxia-inducible factor 1α (HIF1α) and MYC are just as integral to T cell effector generation as nuclear factor of activated T cells (NFAT), activator protein 1 (AP-1) and nuclear factor-κB (NF-κB). Similarly, mammalian target of rapamycin (mTOR) signalling is as crucial in effector T cell activation and differentiation as the activation of mitogen-activated protein kinase (MAPK), protein kinase Cθ (PKCθ) and calcineurin. Thicker arrows indicate the activation of metabolic programmes. Thinner arrows indicate signalling cascades. DAG, diacylglycerol; FOXP3, forkhead box P3; GLUT, glucose transporter; IKK, inhibitor of NF-κB kinase; IL-6R, interleukin-6 receptor; InsP3, inositol-1,4,5-trisphosphate; LAT, linker for activation of T cells; MAPKK, MAPK kinase; PI3K, phosphoinositide 3-kinase; PKCθ, protein kinase Cθ; PLCγ, phospholipase Cγ; PtdInsP2, phosphatidylinositol-4,5-bisphosphate; SLP76, SH2 domain-containing leukocyte protein of 76 kDa (also known as LCP2); STAT, signal transducer and activator of transcription; TCR, T cell receptor; TH cell, T helper cell; ZAP70, ζ-chain-associated protein kinase of 70 kDa.
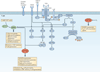
Figure 3. Integrating immunological and metabolic signalling programmes to promote CD8+ memory and CD4+ regulatory T cell generation
This figure depicts the integration of the canonical T cell signalling pathways (blue) and metabolic regulators (green for activated and red for inhibited). AMP-activated protein kinase (AMPK) activation promotes metabolic programmes that enhance the generation of memory and regulatory T (TReg) cells. Alternatively, it is the inhibition of mammalian target of rapamycin (mTOR) and hypoxia-inducible factor 1α (HIF1α) activation that promotes the generation of CD8+ memory or CD4+ regulatory TReg cells. From this perspective, memory T cells and TReg cells share similar metabolic requirements. Thicker arrows indicate the downstream consequences of AMPK activation, and of the inhibition of mTOR and HIF1α. AP-1, activator protein 1; DAG, diacylglycerol; FOXP3, forkhead box P3; IKK, inhibitor of NF-κB kinase; InsP3, inositol-1,4,5-trisphosphate; LAT, linker for activation of T cells; LKB1, liver kinase B1; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; NFAT, nuclear factor of activated T cells; NF-κB, nuclear factor-κB; PI3K, phosphoinositide 3-kinase; PKCθ, protein kinase Cθ; PLCγ, phospholipase Cγ; PtdInsP2, phosphatidylinositol-4,5-bisphosphate; SLP76, SH2 domain-containing leukocyte protein of 76 kDa (also known as LCP2); TCR, T cell receptor; TH cell, T helper cell; ZAP70, ζ-chain-associated protein kinase of 70 kDa.
Similar articles
- mTOR signaling in the differentiation and function of regulatory and effector T cells.
Zeng H, Chi H. Zeng H, et al. Curr Opin Immunol. 2017 Jun;46:103-111. doi: 10.1016/j.coi.2017.04.005. Epub 2017 May 20. Curr Opin Immunol. 2017. PMID: 28535458 Free PMC article. Review. - Natural killer group 2D and CD28 receptors differentially activate mammalian/mechanistic target of rapamycin to alter murine effector CD8+ T-cell differentiation.
McQueen B, Trace K, Whitman E, Bedsworth T, Barber A. McQueen B, et al. Immunology. 2016 Mar;147(3):305-20. doi: 10.1111/imm.12563. Epub 2016 Jan 17. Immunology. 2016. PMID: 26661515 Free PMC article. - The mTOR pathway and integrating immune regulation.
Cobbold SP. Cobbold SP. Immunology. 2013 Dec;140(4):391-8. doi: 10.1111/imm.12162. Immunology. 2013. PMID: 23952610 Free PMC article. Review. - The influence of mTOR on T helper cell differentiation and dendritic cell function.
Salmond RJ, Zamoyska R. Salmond RJ, et al. Eur J Immunol. 2011 Aug;41(8):2137-41. doi: 10.1002/eji.201141523. Epub 2011 Jul 4. Eur J Immunol. 2011. PMID: 21725971 Review. - Metabolism of murine TH 17 cells: Impact on cell fate and function.
Wang R, Solt LA. Wang R, et al. Eur J Immunol. 2016 Apr;46(4):807-16. doi: 10.1002/eji.201545788. Epub 2016 Mar 10. Eur J Immunol. 2016. PMID: 26893133 Free PMC article. Review.
Cited by
- Metabolic signatures of immune cells in chronic kidney disease.
Li J, Yang Y, Wang Y, Li Q, He F. Li J, et al. Expert Rev Mol Med. 2022 Oct 21;24:e40. doi: 10.1017/erm.2022.35. Expert Rev Mol Med. 2022. PMID: 36268748 Free PMC article. Review. - Glucose- and glutamine-fueled stabilization of C-Myc is required for T-cell proliferation and malignant transformation.
Gabriel SS, Kallies A. Gabriel SS, et al. Cell Death Discov. 2016 Jun 27;2:16047. doi: 10.1038/cddiscovery.2016.47. eCollection 2016. Cell Death Discov. 2016. PMID: 27551535 Free PMC article. No abstract available. - Sugar, fat, and protein: new insights into what T cells crave.
Delgoffe GM, Powell JD. Delgoffe GM, et al. Curr Opin Immunol. 2015 Apr;33:49-54. doi: 10.1016/j.coi.2015.01.015. Epub 2015 Feb 6. Curr Opin Immunol. 2015. PMID: 25665466 Free PMC article. Review. - Polyphenols Modulating Effects of PD-L1/PD-1 Checkpoint and EMT-Mediated PD-L1 Overexpression in Breast Cancer.
Messeha SS, Zarmouh NO, Soliman KFA. Messeha SS, et al. Nutrients. 2021 May 19;13(5):1718. doi: 10.3390/nu13051718. Nutrients. 2021. PMID: 34069461 Free PMC article. Review. - ACC1-expressing pathogenic T helper 2 cell populations facilitate lung and skin inflammation in mice.
Nakajima T, Kanno T, Yokoyama S, Sasamoto S, Asou HK, Tumes DJ, Ohara O, Nakayama T, Endo Y. Nakajima T, et al. J Exp Med. 2021 Dec 6;218(12):e20210639. doi: 10.1084/jem.20210639. Epub 2021 Nov 23. J Exp Med. 2021. PMID: 34813654 Free PMC article.
References
- Gerondakis S, Fulford TS, Messina NL, Grumont RJ. NF-κB control of T cell development. Nature Immunol. 2014;15:15–25. - PubMed
- Macian F, et al. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. - PubMed
- Fields PE, Gajewski TF, Fitch FW. Blocked Ras activation in anergic CD4+ T cells. Science. 1996;271:1276–1278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA006973/CA/NCI NIH HHS/United States
- R01 AI091481/AI/NIAID NIH HHS/United States
- T32 AI007247/AI/NIAID NIH HHS/United States
- R01AI077610‑01A2/AI/NIAID NIH HHS/United States
- R01 AI077610/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous