The RAB5-GEF function of RIN1 regulates multiple steps during Listeria monocytogenes infection - PubMed (original) (raw)
. 2014 Nov;15(11):1206-18.
doi: 10.1111/tra.12204. Epub 2014 Sep 4.
Affiliations
- PMID: 25082076
- PMCID: PMC4282165
- DOI: 10.1111/tra.12204
The RAB5-GEF function of RIN1 regulates multiple steps during Listeria monocytogenes infection
Kavitha Balaji et al. Traffic. 2014 Nov.
Erratum in
- The RAB5-GEF Function of RIN1 Regulates Multiple Steps During Listeria monocytogenes Infection.
Balaji K, French CT, Miller JF, Colicelli J. Balaji K, et al. Traffic. 2015 Jul;16(7):796. doi: 10.1111/tra.12268. Epub 2015 Feb 24. Traffic. 2015. PMID: 26050999 No abstract available.
Abstract
Listeria monocytogenes is a food-borne pathogenic bacterium that invades intestinal epithelial cells through a phagocytic pathway that relies on the activation of host cell RAB5 GTPases. Listeria monocytogenes must subsequently inhibit RAB5, however, in order to escape lysosome-mediated destruction. Relatively little is known about upstream RAB5 regulators during L. monocytogenes entry and phagosome escape processes in epithelial cells. Here we identify RIN1, a RAS effector and RAB5-directed guanine nucleotide exchange factor (GEF), as a host cell factor in L. monocytogenes infection. RIN1 is rapidly engaged following L. monocytogenes infection and is required for efficient invasion of intestinal epithelial cells. RIN1-mediated RAB5 activation later facilitates the fusion of phagosomes with lysosomes, promoting clearance of bacteria from the host cell. These results suggest that RIN1 is a host cell regulator that performs counterbalancing functions during early and late stages of L. monocytogenes infection, ultimately favoring pathogen clearance.
Keywords: Listeria monocytogenes; MET; RAB5; RIN1; phagosome.
© 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures
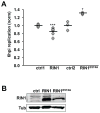
FIGURE 1. RIN1-to-RAB5 signaling is required for efficient L. monocytogenes entry into epithelial cells
A. HeLa cells were infected with L. monocytogenes (500 MOI; 2.5 min) and lysates probed with anti-RIN1pY36, anti-RIN1 and anti-tubulin (Tub). A short time and high MOI were used to detect the transient phosphorylation that occurs upon infection. RIN1pY36 levels were normalized to total RIN1 and tubulin (Figure S1). The ABL tyrosine kinase inhibitor imatinib was used at 5μM. n=2 experiments, p=0.03. B. Control, Rin1 shRNA (Rin1 kd) or RIN1E574A transduced IEC-18 cells were infected at 50 MOI for 1 hour and extracellular bacteria killed using gentamicin. Cell lysate dilutions were plated to determine colony-forming units (CFUs) (ctrl-kd n=17, Rin1-kd n=16, 4 independent experiments, p=1.8×10−16; ctrl-vec and RIN1E574A n=6, 2 independent experiments, p=2.6×10−6). C. Invasion assay (as in B) using IEC-6 Cdx2 cells transduced with control or Rin1-shRNA. Cell lysate dilutions were plated to obtain CFUs (ctrl n=3, Rin1-kd n=6, p=0.003). D. IEC6-Cdx2 cells transduced with control, RIN1 or RIN1E574A vectors were subjected to invasion assays as in B and CFUs determined (ctrl n=9, RIN1 and RIN1E574A n=10, 2 independent experiments, p=2.3×10−6). E. Control or Rin1-shRNA transduced IEC-6 Cdx2 cells were infected (10 MOI) with wild type or InlAmur L. monocytogenes and extracellular bacteria killed using gentamicin. Cell lysate dilutions were plated to obtain CFUs (ctrl n=4, Rin1-kd n=6; wild type L. monocytogenes p=6.7×10−5, InlAmur p=0.002). InlAmur L. monocytogenes invaded more efficiently than wild type (p=0.009). Note: Rin1=rat protein; RIN1=human protein. (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
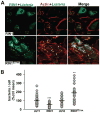
FIGURE 2. RIN1 and RAB5 are recruited to L. monocytogenes during invasion
A and B. IEC-6 Cdx2 cells transduced with RIN1 (A) or RIN1E574A (B) were plated on collagen, infected with L. monocytogenes at MOI 100 for 50 or 80 minutes at 37°C. The cells were washed thoroughly, fixed and stained for RIN1 (green) and RAB5 (red). Host cell nuclei and L. monocytogenes were visualized by DAPI (blue). Arrows show RAB5 and RIN1 recruitment to L. monocytogenes. Insets show magnified versions of the regions pointed by the arrows. Scale bar: 10 μm. C. Quantification of percentage total bacteria co-localizing with RAB5 at each time point in RIN1 and RIN1E574A IEC-6 Cdx2 cells, representative of two independent experiments.
FIGURE 3. RIN1-to-RAB5 signaling inhibits L. monocytogenes replication
A. IEC-18 cells transduced with control, RIN1 or RIN1E574A were infected with L. monocytogenes (50 MOI) after which extracellular bacteria were killed with gentamicin. At 1 hour post-infection (hpi) cells were either lysed or incubated in medium with low gentamicin before lysing at 6 hpi. Dilutions of the lysates were plated to obtain CFUs. Intracellular replication was measured as the fold change in CFU (6 hpi vs. 1 hpi) (ctrl1 and RIN1: n=6, p=1×10−30; ctrl2 and RIN1E574A: n=3, p=0.01). Values are representative of two independent experiments. B. Immunoblot evaluation of RIN1 expression. (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
FIGURE 4. RIN1 inhibits phagosome escape
A. IEC-18 cells transduced with control, RIN1 or RIN1E574A vector were infected with L. monocytogenes (20 MOI) and extracellular bacteria killed with gentamicin, then incubated in medium with low gentamicin for 2 hours. Fixed cells were stained for RIN1 (cyan), actin (red) and L. monocytogenes (green). Scale bar: 10 μm. B. Quantified total bacteria co-localized with actin per cell (actin coating correlates with cytoplasmic localization). Values are representative of two independent experiments and normalized to percent of the control mean. (ctrl1 n=133, RIN1 n=81, p=4 × 10−6, ctrl2 n=70, RIN1E574A n=62, p=0.0012; *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
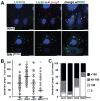
FIGURE 5. RIN1-to-RAB5 signaling promotes lysosome fusion
A. IEC-18 cells transduced with control, RIN1 or RIN1E574A vectors were infected with L. monocytogenes (20 MOI) and extracellular bacteria killed with gentamicin followed by incubation in medium with low gentamicin for 1.5 hours. Fixed cells were stained for RIN1 (cyan), L. monocytogenes (green) and Lamp1 (red). Nuclei were stained with DAPI (blue). Scale bar: 10 μm. B. and C. Quantification of lysosome-localized bacteria in infected IEC-18 cells. B. Each point is the percentage of total bacteria co-localizing with Lamp1 in a given field. C. Each bar is the percent co-localization in distribution categories. Values are representative of two independent experiments and normalized to percent of control mean (ctrl1 n=147, RIN1 n=124, p=9×10−4, ctrl2 n=161, RIN1E574A=105, p=0.03; *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
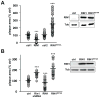
FIGURE 6. RIN1 inhibits bacteria spread between cells
A. IEC-18 cells infected with L. monocytogenes (50 MOI) were trypsinized and transferred to a monolayer of IEC-18 cells transduced with control, RIN1 or RIN1E574A vectors in low gentamicin medium and overlaid with DMEM+0.7% agarose. After 48 hours plaque areas were measured using ImageJ (NIH). Each point is a single plaque area normalized to percent of control mean (ctrl1 n=55, RIN1 n=50, p=3×10−19; ctrl2 n=65, RIN1E574A n=75, p= 2×10−19). Values were obtained from two independent experiments. RIN1 expression confirmed by immunoblot (right). B. Plaque assay as described in A but using IEC-6 Cdx2 cells transduced with control, Rin1-shRNA, RIN1 or RIN1E574A vector (ctrl n=29, Rin1-kd n=21, p=7.1×10−13, RIN1 n=36, p=0.0001, RIN1E574A n=37, p=1.9×10−12). Values were obtained from two independent experiments. Expression confirmed by immunoblot (right). (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005)
FIGURE 7. RIN1 is a RAB5 activator that controls the course of L. monocytogenes infection
InlB binding to MET initiates clathrin-mediated invasion while also triggering activation of RAS and its effectors. RIN1 activates RAB5 by guanine nucleotide exchange. RAB5 facilitates L. monocytogenes internalization (1) but also promotes the fusion of phagosome-encased bacteria with lysosomes (2). This step clears bacteria before its escape and replication in the cytoplasm (3) and blocks spread to adjacent cells (4). Disrupting RIN1 function (silencing or GEF-interfering mutant) lowers invasion efficiency, but impedes the bactericidal response.
Similar articles
- Phagocytosed live Listeria monocytogenes influences Rab5-regulated in vitro phagosome-endosome fusion.
Alvarez-Dominguez C, Barbieri AM, Berón W, Wandinger-Ness A, Stahl PD. Alvarez-Dominguez C, et al. J Biol Chem. 1996 Jun 7;271(23):13834-43. doi: 10.1074/jbc.271.23.13834. J Biol Chem. 1996. PMID: 8662791 - RIN1 orchestrates the activation of RAB5 GTPases and ABL tyrosine kinases to determine the fate of EGFR.
Balaji K, Mooser C, Janson CM, Bliss JM, Hojjat H, Colicelli J. Balaji K, et al. J Cell Sci. 2012 Dec 1;125(Pt 23):5887-96. doi: 10.1242/jcs.113688. Epub 2012 Sep 12. J Cell Sci. 2012. PMID: 22976291 Free PMC article. - Inhibition of early endosome fusion by Rab5-binding defective Ras interference 1 mutants.
Galvis A, Balmaceda V, Giambini H, Conde A, Villasana Z, Fornes MW, Barbieri MA. Galvis A, et al. Arch Biochem Biophys. 2009 Feb;482(1-2):83-95. doi: 10.1016/j.abb.2008.11.009. Epub 2008 Nov 14. Arch Biochem Biophys. 2009. PMID: 19032933 Free PMC article. - Interactions of Listeria monocytogenes with the autophagy system of host cells.
Lam GY, Czuczman MA, Higgins DE, Brumell JH. Lam GY, et al. Adv Immunol. 2012;113:7-18. doi: 10.1016/B978-0-12-394590-7.00008-7. Adv Immunol. 2012. PMID: 22244576 Review. - Role of host GTPases in infection by Listeria monocytogenes.
Ireton K, Rigano LA, Dowd GC. Ireton K, et al. Cell Microbiol. 2014 Sep;16(9):1311-20. doi: 10.1111/cmi.12324. Epub 2014 Aug 4. Cell Microbiol. 2014. PMID: 24948362 Free PMC article. Review.
Cited by
- The complexity of Rab5 to Rab7 transition guarantees specificity of pathogen subversion mechanisms.
Mottola G. Mottola G. Front Cell Infect Microbiol. 2014 Dec 22;4:180. doi: 10.3389/fcimb.2014.00180. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25566515 Free PMC article. Review. No abstract available. - Role of Rab GTPases in Bacteria Escaping from Vesicle Trafficking of Host Cells.
Xu H, Wang S, Wang X, Zhang P, Zheng Q, Qi C, Liu X, Li M, Liu Y, Liu J. Xu H, et al. J Microbiol. 2024 Aug;62(8):581-590. doi: 10.1007/s12275-024-00162-9. Epub 2024 Aug 30. J Microbiol. 2024. PMID: 39212865 Review. - Activation of Mitofusin2 by Smad2-RIN1 Complex during Mitochondrial Fusion.
Kumar S, Pan CC, Shah N, Wheeler SE, Hoyt KR, Hempel N, Mythreye K, Lee NY. Kumar S, et al. Mol Cell. 2016 May 19;62(4):520-31. doi: 10.1016/j.molcel.2016.04.010. Epub 2016 May 12. Mol Cell. 2016. PMID: 27184078 Free PMC article.
References
- Humphries AC, Way M. The non-canonical roles of clathrin and actin in pathogen internalization, egress and spread. Nature reviews Microbiology. 2013;11(8):551–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA136699/CA/NCI NIH HHS/United States
- CA136699/CA/NCI NIH HHS/United States
- U54 A1065359/PHS HHS/United States
- P30 DK041301/DK/NIDDK NIH HHS/United States
- T32 GM007185/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous