mTOR signaling regulates nucleolar targeting of the SUMO-specific isopeptidase SENP3 - PubMed (original) (raw)
mTOR signaling regulates nucleolar targeting of the SUMO-specific isopeptidase SENP3
Nithya Raman et al. Mol Cell Biol. 2014 Dec.
Abstract
Ribosome biogenesis is a multistep cellular pathway that involves more than 200 regulatory components to ultimately generate translation-competent 80S ribosomes. The initial steps of this process, particularly rRNA processing, take place in the nucleolus, while later stages occur in the nucleoplasm and cytoplasm. One critical factor of 28S rRNA maturation is the SUMO-isopeptidase SENP3. SENP3 tightly interacts with the nucleolar scaffold protein NPM1 and is associated with nucleolar 60S preribosomes. A central question is how changes in energy supply feed into the regulation of ribosome maturation. Here, we show that the nutrient-sensing mTOR kinase pathway controls the nucleolar targeting of SENP3 by regulating its interaction with NPM1. We define an N-terminal domain in SENP3 as the critical NPM1 binding region and provide evidence that mTOR-mediated phosphorylation of serine/threonine residues within this region fosters the interaction of SENP3 with NPM1. The inhibition of mTOR triggers the nucleolar release of SENP3, thereby likely compromising its activity in rRNA processing. Since mTOR activity is tightly coupled to nutrient availability, we propose that this pathway contributes to the adaptation of ribosome maturation in response to the cellular energy status.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
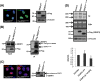
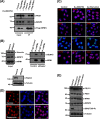
FIG 1
Nucleolar SENP3 is required for ribosome biogenesis. (A, left) HeLa cells transiently expressing Flag-SENP3 or Flag-SENP3Δ76-159 were stained with anti-Flag antibody to determine their localization. (Right) Immunoblotting of the transfected samples was performed with anti-Flag antibody to test for their expression levels, and antitubulin was used as a loading control. (B) Flag-tagged SENP3Δ76-159 was expressed in HEK293T cells and purified by immunoprecipitation (IP) with anti-Flag beads. Flag-SENP3Δ76-159 along with the interacting proteins was eluted from the beads with SDS sample buffer, separated by SDS-PAGE, and detected using anti-Flag or anti-SENP3 antibody, respectively. (C) As described for panel A, except the cells were stained with anti-SENP3 antibody to simultaneously determine the localization of exogenous and endogenous SENP3 (left), and the level of the endogenous SENP3 was compared with the exogenous SENP3 levels by immunoblotting using anti-SENP3 antibody (right). (D) HeLa cells were transiently transfected with wild-type SENP3 or the SENP3Δ76-159 variant. After 48 h, the cells were pulse labeled with [32P]orthophosphate for 1 h and chased for 1 h with unlabeled medium. RNA was purified from cells, separated under denaturing conditions on an agarose gel, and subjected to autoradiography. (Upper) The steady-state RNA levels were visualized by ethidium bromide (EtBr) staining of the gel. Transfection efficiency was monitored by immunoblotting using an anti-Flag antibody. Antivinculin staining served as a loading control. (Lower) The 28/32S rRNA ratio then was quantified using a phosphorimager. Values represent averages from three independent experiments, with error bars indicating standard deviations (SD), and a P value of <0.05.
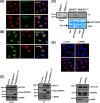
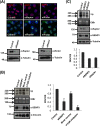
FIG 2
NPM1 interaction is required for the nucleolar targeting of SENP3. (A) HeLa cells transiently expressing Flag-SENP3 or Flag-SENP3Δ76-159 were stained with anti-Flag and anti-PELP1 antibodies to determine their localization. (B) As described for panel A, except the cells were stained with anti-Flag and anti-NPM1 antibodies after transfection. (C) HeLa cells were transiently transfected with Flag-SENP3 or Flag-SENP3Δ76-159, and their interaction with NPM1 and PELP1 was determined after immunoprecipitation on Flag beads by immunoblotting using anti-Flag, anti-NPM1, and anti-PELP1 antibodies. (D) [35S]methionine-labeled SENP3 and SENP3Δ76-159 were generated by in vitro transcription/translation and used for GST pulldown assays using GST or GST-NPM1 bound to glutathione-Sepharose beads. After separation by SDS-PAGE, interactions were detected via autoradiography. (Upper) Autoradiography showing 35S-labeled SENP3 variants and their interaction with GST-NPM1. (Lower) Coomassie staining of the SDS-PAGE gel showing GST and GST-NPM1. (E) HeLa cells were either mock transfected or transfected with siRNA directed against NPM1. Seventy-two hours posttransfection, the cells were fixed and stained with anti-SENP3 or anti-NPM1 antibody in order to visualize their respective localizations through indirect immunofluorescence (top). Western blotting was performed with anti-SENP3 and anti-NPM1 antibodies to check for their proteins levels, and antivinculin was used as a loading control (bottom).
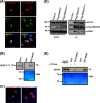
FIG 3
Localization and NPM1 interaction of SENP3 is determined by its N-terminal serine/threonine residues. (A) Full-length Flag-SENP3 and Flag-SENP31-195 were transiently expressed in HeLa cells, and their colocalization with NPM1 was determined by indirect immunofluorescence using anti-Flag and anti-NPM1 antibodies, respectively. (B) [35S]methionine-labeled SENP31-195 was generated by in vitro transcription/translation and tested for NPM1 interaction by GST pulldown using GST-NPM1 as the bait. CBB, Coomassie brilliant blue. (C) HeLa cells were transiently transfected with plasmids expressing wild-type Flag-SENP3 or Flag-SENP328S→A and stained with anti-Flag antibody for indirect immunofluorescence. (D) Interaction of wild-type Flag-SENP3 and Flag-SENP328S→A with endogenous NPM1 and PELP1 was monitored by Western blotting after immunoprecipitation of the transiently expressed Flag-tagged proteins in HeLa cells. The different lanes shown originate from the same blot taken at the same exposure times. (E) [35S]methionine-labeled SENP3 was generated by in vitro transcription/translation and either mock treated or treated with lambda phosphatase (λ-PPase). The proteins were used for a pulldown assay with either GST or GST-NPM1, and the interaction of SENP3 with NPM1 was detected by autoradiography.
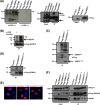
FIG 4
mTOR phosphorylates SENP3 at its N terminus. (A) HEK293T cells were transfected with plasmids expressing Flag-SENP3, Flag-NPM1, Flag-4EBP1, or empty vector, and the respective proteins were immunoprecipitated using Flag-agarose beads. The proteins bound to the beads then were used for in vitro phosphorylation assay in the presence of [32P]ATP with or without the catalytic fragment of mTOR (aa 1362 to 2549). (Left) Subsequently, the samples were separated by SDS-PAGE and phosphorylation was detected by autoradiography. IgGs are indicated by an asterisk. (Right) The expression and immunoprecipitation of the Flag-tagged constructs was monitored by Western blotting with anti-Flag antibody. (B) Flag-tagged SENP3 was immunoprecipitated from HeLa cells and used for in vitro phosphorylation assay with [32P]ATP with either the mock-treated or Torin1-treated mTOR/Raptor/MLST8 complex. (Upper) The samples then were subjected to SDS-PAGE, followed by autoradiography. mTOR autophosphorylation is indicated by an asterisk. (Lower) Immunopurified SENP3 was monitored by Western blotting with anti-Flag antibody. (C, upper) mTOR-mediated phosphorylation of Flag-SENP3 or Flag-SENP31-195 was analyzed in an in vitro phosphorylation assay as described for panel A. (Lower) IP was verified by immunoblotting with anti-Flag antibody. (D) mTOR-mediated phosphorylation of wild-type Flag-SENP3 and Flag-SENP328S→A was done as described for panels A and B. (Lower) IPs were verified by immunoblotting with anti-Flag antibody. (E) HeLa cells were transiently transfected with plasmids expressing wild-type Flag-SENP3, Flag-SENP37S→A, or Flag-SENP36A→S and stained with anti-Flag antibody for indirect immunofluorescence. (F) Interaction of wild-type Flag-SENP3, Flag-SENP37S→A, Flag-SENP36A→S, or Flag-SENP328S→A with endogenous NPM1 and PELP1 was monitored by Western blotting after immunoprecipitation of the transiently expressed Flag-tagged proteins in HeLa cells.
FIG 5
mTOR affects SENP3 localization and SENP3-NPM1 interaction. (A) U2OS cells expressing Flag-NPM1 were either mock treated or treated with the mTOR inhibitor Ku-0063794. Subsequently, Flag-NPM1 was immunoprecipitated from these cells using Flag-agarose beads and separated by SDS-PAGE. Anti-Flag antibody was used to verify the immunoprecipitation of Flag-NPM1, and anti-SENP3 or anti-PES1 antibody was used to check for coimmunoprecipitation. (B) HeLa cells were transfected with either control siRNA or siRNA directed against Raptor, and 72 h later the cells were harvested for immunoprecipitation of the endogenous NPM1. (Upper right) The samples then were probed by immunoblotting with anti-NPM1 or anti-SENP3 antibody to check for NPM1 levels and coimmunoprecipitation of SENP3. (Upper left and lower) The proteins levels in control and knockdown samples were monitored by Western blotting with anti-NPM1, anti-SENP3, and anti-Raptor antibodies with antitubulin and antivinculin antibodies as loading controls. (C) HeLa cells were either mock treated, treated with Ku-0063794, or starved in Earle's balanced salt solution (EBSS) for 6 h and stained with anti-SENP3, anti-NPM1, anti-PES1, and anti-PELP1 antibodies to visualize their respective localization by indirect immunofluorescence. (D) SENP3 localization in HeLa cells was detected by immunofluorescence using anti-SENP3 antibody after treatment with rapamycin. (E) Immunoblotting of the HeLa cell lysates was done to check for changes in protein levels using anti-PELP1, anti-SENP3, anti-PES1, anti-S6K(T389-P), and anti-NPM1 antibodies after EBSS starvation or rapamycin or Ku-0063794 treatment. Antitubulin antibody was used as a loading control.
FIG 6
Inhibition of mTOR signaling reduces 28S maturation. (A, upper) The localization of SENP3 and NPM1 was monitored by immunofluorescence in HeLa cells after the siRNA-mediated depletion of Rictor or Raptor. (Lower) Efficient depletion of the respective proteins was verified by immunoblotting with anti-Raptor or anti-Rictor antibody with antitubulin serving as a loading control. (B) A pulse-chase rRNA processing assay, as described in the legend to Fig. 1D, was performed in HeLa cells after siRNA-mediated depletion of SENP3 or treatment of cells with Ku-0063794. After extraction, the RNA was run on a denaturing agarose gel, stained with EtBr, and subjected to autoradiography for the detection of the various rRNA species. The efficiency of depletion and levels of SENP3 was controlled by Western blotting with anti-SENP3 antibody. Vinculin was used as a loading control. The 28/32S rRNA ratio was quantified using a phosphorimager. The values represent averages from 3 independent experiments, and the error bars indicate standard errors of the means. (C) As described for panel B, except the cells first were transfected with control siRNA or siRNA directed against SENP3 or Raptor before the pulse-chase assay. The efficient depletion of the respective proteins was monitored by immunoblotting with anti-SENP3 and anti-Raptor antibodies, using tubulin as a loading control. Phosphorimager quantifications are as shown in the lower panel.
Similar articles
- The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing.
Haindl M, Harasim T, Eick D, Muller S. Haindl M, et al. EMBO Rep. 2008 Mar;9(3):273-9. doi: 10.1038/embor.2008.3. Epub 2008 Feb 8. EMBO Rep. 2008. PMID: 18259216 Free PMC article. - The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex.
Finkbeiner E, Haindl M, Muller S. Finkbeiner E, et al. EMBO J. 2011 Mar 16;30(6):1067-78. doi: 10.1038/emboj.2011.33. Epub 2011 Feb 15. EMBO J. 2011. PMID: 21326211 Free PMC article. - The AAA ATPase MDN1 Acts as a SUMO-Targeted Regulator in Mammalian Pre-ribosome Remodeling.
Raman N, Weir E, Müller S. Raman N, et al. Mol Cell. 2016 Nov 3;64(3):607-615. doi: 10.1016/j.molcel.2016.09.039. Mol Cell. 2016. PMID: 27814492 - SUMO routes ribosome maturation.
Finkbeiner E, Haindl M, Raman N, Muller S. Finkbeiner E, et al. Nucleus. 2011 Nov-Dec;2(6):527-32. doi: 10.4161/nucl.2.6.17604. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064470 Review. - Signal Transduction in Ribosome Biogenesis: A Recipe to Avoid Disaster.
Piazzi M, Bavelloni A, Gallo A, Faenza I, Blalock WL. Piazzi M, et al. Int J Mol Sci. 2019 Jun 3;20(11):2718. doi: 10.3390/ijms20112718. Int J Mol Sci. 2019. PMID: 31163577 Free PMC article. Review.
Cited by
- Brucella effectors NyxA and NyxB target SENP3 to modulate the subcellular localisation of nucleolar proteins.
Louche A, Blanco A, Lacerda TLS, Cancade-Veyre L, Lionnet C, Bergé C, Rolando M, Lembo F, Borg JP, Buchrieser C, Nagahama M, Gérard FCA, Gorvel JP, Gueguen-Chaignon V, Terradot L, Salcedo SP. Louche A, et al. Nat Commun. 2023 Jan 6;14(1):102. doi: 10.1038/s41467-022-35763-8. Nat Commun. 2023. PMID: 36609656 Free PMC article. - SUMOylation- and GAR1-Dependent Regulation of Dyskerin Nuclear and Subnuclear Localization.
MacNeil DE, Lambert-Lanteigne P, Qin J, McManus FP, Bonneil E, Thibault P, Autexier C. MacNeil DE, et al. Mol Cell Biol. 2021 Mar 24;41(4):e00464-20. doi: 10.1128/MCB.00464-20. Print 2021 Mar 24. Mol Cell Biol. 2021. PMID: 33526451 Free PMC article. - Hypoxia-driven deSUMOylation of EXOSC10 promotes adaptive changes in the transcriptome profile.
Filippopoulou C, Thomé CC, Perdikari S, Ntini E, Simos G, Bohnsack KE, Chachami G. Filippopoulou C, et al. Cell Mol Life Sci. 2024 Jan 27;81(1):58. doi: 10.1007/s00018-023-05035-9. Cell Mol Life Sci. 2024. PMID: 38279024 Free PMC article. - SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes.
Zhao X. Zhao X. Mol Cell. 2018 Aug 2;71(3):409-418. doi: 10.1016/j.molcel.2018.07.027. Mol Cell. 2018. PMID: 30075142 Free PMC article. Review. - Structural basis for the human SENP5's SUMO isoform discrimination.
Sánchez-Alba L, Ying L, Maletic MD, De Bolòs A, Borràs-Gas H, Liu B, Varejão N, Amador V, Mulder MPC, Reverter D. Sánchez-Alba L, et al. Nat Commun. 2025 May 22;16(1):4764. doi: 10.1038/s41467-025-60029-4. Nat Commun. 2025. PMID: 40404649 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous