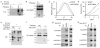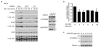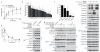SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1 - PubMed (original) (raw)
. 2015 Mar 26;519(7544):477-81.
doi: 10.1038/nature14107. Epub 2015 Jan 7.
Lorena Pochini 2, Taras Stasyk 3, Mariana E G de Araújo 3, Michele Galluccio 2, Richard K Kandasamy 1, Berend Snijder 1, Astrid Fauster 1, Elena L Rudashevskaya 1, Manuela Bruckner 1, Stefania Scorzoni 1, Przemyslaw A Filipek 3, Kilian V M Huber 1, Johannes W Bigenzahn 1, Leonhard X Heinz 1, Claudine Kraft 4, Keiryn L Bennett 1, Cesare Indiveri 2, Lukas A Huber 3, Giulio Superti-Furga 1
Affiliations
- PMID: 25561175
- PMCID: PMC4376665
- DOI: 10.1038/nature14107
SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1
Manuele Rebsamen et al. Nature. 2015.
Abstract
Cell growth and proliferation are tightly linked to nutrient availability. The mechanistic target of rapamycin complex 1 (mTORC1) integrates the presence of growth factors, energy levels, glucose and amino acids to modulate metabolic status and cellular responses. mTORC1 is activated at the surface of lysosomes by the RAG GTPases and the Ragulator complex through a not fully understood mechanism monitoring amino acid availability in the lysosomal lumen and involving the vacuolar H(+)-ATPase. Here we describe the uncharacterized human member 9 of the solute carrier family 38 (SLC38A9) as a lysosomal membrane-resident protein competent in amino acid transport. Extensive functional proteomic analysis established SLC38A9 as an integral part of the Ragulator-RAG GTPases machinery. Gain of SLC38A9 function rendered cells resistant to amino acid withdrawal, whereas loss of SLC38A9 expression impaired amino-acid-induced mTORC1 activation. Thus SLC38A9 is a physical and functional component of the amino acid sensing machinery that controls the activation of mTOR.
Figures
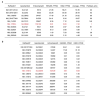
Extended Data 1. Expression of SLC members of amino acid transporter families
a, Table of SLCs belonging to amino acid transporter families robustly expressed in HEK293 and K562 cells as monitored by RNAseq. SLC members of amino acid transporter-containing families (SLC1, 6, 7, 16, 17, 18, 32, 36, 38 and 43 families) expressed (FPKM>0.5) in both cell lines were ranked according to their expression level, top ten are shown. The number of PubMed entries was obtained by querying the GeneSymbol (24th October 2013). b, Expression of members of the SLC32, SLC36 and SLC38 families in HEK293 and K562 cells.
Extended Data 2. Biochemical and functional characterisation of SLC38A9
a-b, Where indicated, HEK293T cells were transfected with the tagged SLC38A9 constructs (+) or empty vector (−). Cell lysates were left untreated (Untr.) or incubated 1 hour at 37 °C in presence or absence of PNGase and analysed by immunoblot. Results are representative of two independent experiments (n=2) c, Cell size measurements of HEK293T cells after short-hairpin (shRNA) mediated knockdown against GFP (control, dashed black line) or SLC38A9 (grey line), measured by automated microscopy and image analysis. Sparse and interphase cells were selected using image analysis and machine learning, and nucleus diameter was used as robust proxy for cell size. Smoothed distributions of 2400 and 4165 cells, respectively, are shown. d, Cell proliferation measurement of HEK293T cells transduced with lentivirus-encoded shRNA against SLC38A9 or GFP. 105 cells were seeded and counted every 24 h. Mean values ± s.d. from triplicates. Results are representative of two independent experiments (n=2). e-f, Where indicated, HEK293T cells were transfected with the tagged SLC38A9. Cell lysates were prepared and left untreated (Untr.) or incubated 1 hour at 37 °C in presence or absence of PNGase and analysed by immunoblot, Where indicated, cell lysates were boiled for 5 min at 95 °C after PNGase treatment. g-h, Lysates from murine NIH/3T3 (g) or Raw 264.7 (h) cells were subjected to immunoprecipitation with the indicated antibodies, treated with PNGase and analysed by immunoblot. Results are representative of two independent experiments (n=2). <: SLC38A9; *: non-specific band.
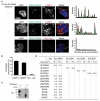
Extended Data 3. SLC38A9 proteomic analysis: bait localisation and results
a, Single-channel and merged confocal microscopy images of DAPI stained nuclei and indirect immunofluorescence against HA-tagged SLC38A9 and endogenous lysosomal markers LAMP1 (top panel) and LAMP2 (middle panel) and the non-induced and secondary-antibody only control (bottom panel) in HEK293 Flp-In TREx cells. Scale bar, 10 μm. Intensity profiles for SLC38A9 (green) and LAMP1, LAMP2 or secondary antibody control (red) along the cross-section lines indicated in the respective merged channel images are shown. b, Quantification of HA-SLC38A9 signal above background (dashed lines in a) that colocalizes with LAMP1, LAMP2 or secondary-antibody only positive areas. Average and s.d. of at least two images is shown, analysing colocalization in 22, 34 and 27 cells respectively. c, HEK293 Flp-In TREx cells inducibly expressing SLC38A9 were treated or not with doxycycline (Dox) for 24h. Where indicated, cell lysates were treated with PNGase and analysed by immunoblot. d, Tabular view summarizing the proteomic analysis of SLC38A9, SLC38A1, SLC38A2 and SLC36A1. Comparison of the SLC38A9 interactors identified by TAP–LC-MS/MS to the same analysis performed with the other transporters. Spectral counts (Sp. c., average of biological replicates) and sequence coverage (Sq. c., percentage, average of biological replicates) are indicated. Data shown are based on two independent experiments for each condition (n=2), each analysed in two technical replicates.
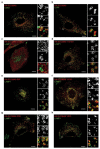
Extended Data 4. SLC38A9 localizes to the late endosome/lysosome compartment
a-h, HeLa cells were transfected with the indicated strep-HA tagged SLC38A9 construct. Merged and single-channel confocal microscopy images of indirect immunofluorescence of HA-tagged SLC38A9 (red) and endogenous lysosomal marker LAMP1 (green) are shown. Representative cells are shown. Scale bar, 10 μm.
Extended Data 5. SLC38A9 is an integral component of the Ragulator/RAG GTPase complex
a, Tabular view of spectral counts (Spec. count, average of biological replicates) and sequence coverage (Seq. cov., percentage, average of biological replicates) of the core Ragulator/RAG GTPase network and published interactors detected. Data shown are based on two independent experiments for each condition (n=2), and analysed in two technical replicates b-c, SLC38A9 peptides detected in LAMTOR1, 3, 4 and 5 (b) or in SLC38A9 (c) TAP-MS/MS analysis are mapped on SLC38A9 sequence and highlighted in bold. Transmembrane helices are highlighted in light brown. Potential tryptic cleavage sites are in red.
Extended Data 6. The cytoplasmic N-terminal region of SLC38A9 binds the Ragulator/RAG GTPase complex through evolutionary conserved motifs
a, Sequence alignment of the N-terminal cytoplasmic region (amino acids 1-112) of human, mouse, rat, Xenopus and zebrafish SLC38A9. Amino acids selected for deletion and motifs substituted to alanine are highlighted. Black and grey shading indicates > 60% amino acid sequence identity and similarity, respectively. b-c, HEK293T cells were transfected with the indicated tagged SLC38A9 constructs. Anti-HA immunoprecipitates and cell extracts were analysed by immunoblot. SLC38A9 mutant constructs are labelled with the number of the encoded amino acids (b) or with the amino acid motif substituted to alanine (c). Results are representative of two independent experiments (n=2).
Extended Data 7. Characterization of SLC38A9-mediated amino acid transport in proteoliposomes
a, Purification of SLC38A9. Lanes represent empty vector control and SLC38A9 expressed in E. coli and purified on Ni-chelating chromatography. Immunoblot of the same fractions using anti-His antibody or anti-SLC38A9 are shown. b, Orientation of SLC38A9 in proteoliposomes. Purified His-SLC38A9 protein or proteoliposomes reconstituted with SLC38A9 were incubated overnight at 37°C in presence or in absence of 1 U thrombin. Proteoliposomes were then solubilized with SDS and analysed by immunoblot. Results are representative of two independent experiments (n=2) c, Time course of glutamine uptake by SLC38A9 in proteoliposomes reconstituted with the purified protein fraction. The uptake of 10 μM [3H]-glutamine was measured at different time intervals in the presence of the indicated intraliposomal sodium concentrations. Transport was calculated by subtracting the radioactivity associated to proteoliposomes reconstituted with the empty vector fraction. Values represent means of specific transport ± s.d. from three independent experiments (n=3) d, Time course of glutamine uptake in proteoliposomes reconstituted with purified SLC38A9 wild-type or N128A mutant protein. Values represent means of specific transport ± s.d. from 3 independent experiments (n=3). Significance was estimated by Student’s t test (* P < 0.01). Immunoblot analysis of purified protein reconstituted in the proteoliposomes e. Effect of pH on the reconstituted SLC38A9. Reconstitution and transport assay were performed at the indicated pH. Results are means of specific transport rate ± s.d. from three different experiments (n=3). f, Inhibition of the [3H]-glutamine uptake in proteoliposomes. 1 mM MeAIB (α-(methylamino)isobutyric acid) was added together with 10 μM [3H]-glutamine. Transport was measured at 60 min. Values represent means of percent residual activity with respect to control (without added inhibitor) ± s.d. from three independent experiments (n=3).
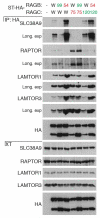
Extended Data 8. Nucleotide-loading/conformation dependent interaction of RAGB/RAGC heterodimers to SLC38A9
HEK293T cells were transfected with the indicated combination of tagged RAG GTPases mutant constructs or empty vector (−). Anti-HA immunoprecipitates and cell extracts were treated with PNGase and analysed by immunoblot. W: wild type; 75: S75N; 120: Q120L; 99: Q99L; 54: T54N. Results are representative of two independent experiments (n=2).
Extended Data 9. Stable expression of SLC38A9 mediates sustained mTORC1 activation upon amino acid starvation
a. SLC38A9- or METAP2-stably expressing HEK293T cells were starved for the indicated time in medium without amino acids and serum. Cell lysates were analysed by immunoblot. Results are representative of two independent experiments (n=2) b. Representative images in the GFP channels of HEK293T cells stably expressing EGFP-LC3B and SLC38A9 or METAP2 starved for 120 min (related to Fig 4 B). Scale bar, 40 μm c. HEK293T cells stably expressing TFEB-STHA and SLC38A9 or METAP2 were starved for the indicated time. Cytoplasmic and nuclear fraction were analysed by immunoblot. Results are representative of two independent experiments (n=2). d. Immunoblot analysis of HEK293T cells stably expressing the indicated SLC38A9 constructs. e. SLC38A9 (S)- or METAP2 (M)-stably expressing HEK293T were starved for 50 min and then stimulated with amino acids for 20 min. Where indicated, cells were treated with ConcanamycinA (5μM) or DMSO during both incubation time. Cell lysates were analysed by immunoblot with the indicated antibodies. Results are representative of two independent experiments (n=2) f. SLC38A9 (S)- or METAP2 (M)-stably expressing HEK293T were treated for 30 min with DMSO (D), ConcanamycinA (C, 5μM) or Torin 1 (T, 250 nM) and then starved for the indicated times in presence of the inhibitors. Cell lysates were analysed by immunoblot. Results are representative of two independent experiments (n=2).
Extended Data 10. Expression of SLC38A9 is required for amino acid-induced mTORC1 activation and is not affected by starvation
a. HeLa were transfected with siRNA targeting SLC38A9 (SLC), LAMTOR1 (LT1) or non targeting control (CNTR). After 72h, cells were starved for 50 min in medium without amino acids and serum and then stimulated with amino acids in presence of insulin (1 μM). Cell lysates were analysed by immunoblot. Results are representative of three independent experiments (n=3). b-c, HEK293T cells were starved for the indicated times. SLC38A9 expression was analysed by quantitative PCR (b) and immunoblot (c). In (b) mean values ± s.d. from technical triplicates is shown. Results are representative of two independent experiments (n=2) <: SLC38A9; *: non-specific band.
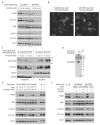
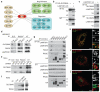
Figure 1. SLC38A9 is a lysosomal component of the amino acid-sensing machinery controlling mTORC1
a, Interactors of SLC38A9 identified by TAP–LC-MS/MS. Data shown are based on two independent experiments (n=2), each analysed in two technical replicates. b-g, Lysates from HEK293T cells transfected with the indicated tagged constructs or empty vector (−) (b, c, g), control (empty vector or shGFP) or shSLC38A9 transduced HEK293T (d) and HeLa (e), or K562 (f) cells were subjected to immunoprecipitation. PNGase-treated immunoprecipitates (IP) and protein extracts (XT) analysed by immunoblot with the indicated antibodies. Results are representative of two independent experiments (n=2) <: ST-HA-SLC38A9; *: non-specific band h-j, Confocal microscopy images of HeLa cells transfected with tagged SLC38A9 construct and immunostained with the anti-HA and LAMP1(h), EEA1(i) or Giantin (j) antibodies. Representative cells are shown. Scale bar, 10 μm.
Figure 2. SLC38A9 is an integral component of the Ragulator/RAG GTPases machinery
a, Interactors of LAMTOR1, LAMTOR3, LAMTOR4, LAMTOR5, RAGA and RAGC were identified by TAP–LC-MS/MS. Proteins that interacted with all the bait proteins are shown, with the addition of RAGD that was not detected in RAGC pulldown. Previously published interactors of the Ragulator/RAG GTPases complex detected are indicated. Data shown are based on two independent experiments for each condition (n=2), each analysed in two technical replicates b-e, HEK293T cells were transfected with the indicated tagged constructs or empty vector (−). Immunoprecipitates and cell extracts were treated with PNGase and analysed by immunoblot. SLC38A9 mutant constructs are labelled with the number of the encoded amino acids (d) or with the amino acid motif substituted to alanine (e). Results are representative of two independent experiments (n=2).
Figure 3. SLC38A9 transport amino acids and interacts with the RAG GTPases in a nucleotide loading- and amino acid sensitive-manner
a, Time course of [3H]-glutamine uptake in proteoliposomes reconstituted with purified SLC38A9 or with control empty vector. Values represent means of specific transport ± s.d. from eight different experiments (n=8). b, Inhibition by amino acids of [3H]-glutamine uptake in proteoliposomes. Values represent means of residual activity with respect to control (without added competitor) ± s.d. from three independent experiments (n=3). c, Uptake of the indicated [3H]-labelled amino acids by SLC38A9 in proteoliposomes. Values represent means of % respect to glutamine transport measured in the same experiment ± s.d. from three independent experiments (n=3). d, Time course of glutamine efflux from proteoliposomes reconstituted with SLC38A9. Values represent means of specific transport ± s.d. from three independent experiments (n=3). e, HEK293T cells were transfected with the indicated combination of tagged RAG GTPases mutant constructs or empty vector (−). PNGase-treated immunoprecipitates and cell extracts were analysed by immunoblot. W: wild type; 66: Q66L; 21: T21N; 75: S75N; 120: Q120L. f-g, HEK293T cells stably expressing the indicated constructs were starved for amino acids and serum for 50 min (AA starv +) and stimulated with amino acids for 20 min (AA stim +). Immunoprecipitates and cell extracts were analysed by immunoblot. In g results of two biological replicates are shown. *: IgG light chain. Results are representative of two (e-f, n=2) or three (g, n=3) independent experiments. In a-c significance was estimated by Student’s t test (* P < 0.01 or ** P < 0.001)
Figure 4. SLC38A9 is a positive regulator of mTORC1 required for its activation by amino acids
a, Wild-type, FLAG-SLC38A9- or FLAG-METAP2-stably expressing HEK293T cells were starved for 30 min in medium without amino acids and serum. Cell lysates were analysed by immunoblot b, HEK293T cells stably expressing EGFP-LC3B and SLC38A9 or METAP2 were starved for the indicated time. LC3B positive autophagosomes were quantified by image analysis. Data were normalized to cell size and plotted relative to the fitted METAP2 maximum. Mean ± s.d of at least three replicate wells. c. HEK293T cells stably expressing the indicated untagged SLC38A9 constructs were treated and analysed as in a. d-e, HEK293T cells transduced with lentivirus-encoded shRNA against SLC38A9 or GFP were starved for 50 min and then stimulated with amino acids (d) or cycloheximide (e, 25μg/ml) for 10 or 20 min. Cell lysates were analysed by immunoblot. f, HEK293T were transfected with siRNA targeting SLC38A9, LAMTOR1 or non targeting control. After 72h, cells were treated as in d. In (e-f lower**)** cell lysates were treated with PNGase**;** *: non-specific band. g, Schematic model of SLC38A9 function in amino acid-induced mTORC1 activation. Results are representative of two (a-e, n=2) or three (f) independent experiments (n=3).
Similar articles
- Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9.
Jung J, Genau HM, Behrends C. Jung J, et al. Mol Cell Biol. 2015 Jul;35(14):2479-94. doi: 10.1128/MCB.00125-15. Epub 2015 May 11. Mol Cell Biol. 2015. PMID: 25963655 Free PMC article. - SLC38A9: A lysosomal amino acid transporter at the core of the amino acid-sensing machinery that controls MTORC1.
Rebsamen M, Superti-Furga G. Rebsamen M, et al. Autophagy. 2016 Jun 2;12(6):1061-2. doi: 10.1080/15548627.2015.1091143. Epub 2015 Oct 2. Autophagy. 2016. PMID: 26431368 Free PMC article. - The amino acid transporter SLC38A9 regulates MTORC1 and autophagy.
Jin M, Klionsky DJ. Jin M, et al. Autophagy. 2015;11(10):1709-10. doi: 10.1080/15548627.2015.1084461. Autophagy. 2015. PMID: 26506891 Free PMC article. - Amino acid signalling upstream of mTOR.
Jewell JL, Russell RC, Guan KL. Jewell JL, et al. Nat Rev Mol Cell Biol. 2013 Mar;14(3):133-9. doi: 10.1038/nrm3522. Epub 2013 Jan 30. Nat Rev Mol Cell Biol. 2013. PMID: 23361334 Free PMC article. Review. - Amino acids and mTORC1: from lysosomes to disease.
Efeyan A, Zoncu R, Sabatini DM. Efeyan A, et al. Trends Mol Med. 2012 Sep;18(9):524-33. doi: 10.1016/j.molmed.2012.05.007. Epub 2012 Jun 28. Trends Mol Med. 2012. PMID: 22749019 Free PMC article. Review.
Cited by
- L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia.
Wang Q, Holst J. Wang Q, et al. Am J Cancer Res. 2015 Mar 15;5(4):1281-94. eCollection 2015. Am J Cancer Res. 2015. PMID: 26101697 Free PMC article. Review. - Mobilization of cholesterol induces the transition from quiescence to growth in Caenorhabditis elegans through steroid hormone and mTOR signaling.
Schmeisser K, Kaptan D, Raghuraman BK, Shevchenko A, Rodenfels J, Penkov S, Kurzchalia TV. Schmeisser K, et al. Commun Biol. 2024 Jan 24;7(1):121. doi: 10.1038/s42003-024-05804-7. Commun Biol. 2024. PMID: 38267699 Free PMC article. - Beyond indigestion: emerging roles for lysosome-based signaling in human disease.
Ferguson SM. Ferguson SM. Curr Opin Cell Biol. 2015 Aug;35:59-68. doi: 10.1016/j.ceb.2015.04.014. Epub 2015 May 15. Curr Opin Cell Biol. 2015. PMID: 25950843 Free PMC article. Review. - Effect of the lysosomotropic agent chloroquine on mTORC1 activation and protein synthesis in human skeletal muscle.
Borack MS, Dickinson JM, Fry CS, Reidy PT, Markofski MM, Deer RR, Jennings K, Volpi E, Rasmussen BB. Borack MS, et al. Nutr Metab (Lond). 2021 Jun 12;18(1):61. doi: 10.1186/s12986-021-00585-w. Nutr Metab (Lond). 2021. PMID: 34118944 Free PMC article. - Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes.
Takahara T, Amemiya Y, Sugiyama R, Maki M, Shibata H. Takahara T, et al. J Biomed Sci. 2020 Aug 17;27(1):87. doi: 10.1186/s12929-020-00679-2. J Biomed Sci. 2020. PMID: 32799865 Free PMC article. Review.
References
Main text references
- Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Current opinion in genetics & development. 2013;23:53–62. doi:10.1016/j.gde.2012.12.005. - PubMed
Method references
- Varjosalo M, et al. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nature methods. 2013;10:307–314. doi:10.1038/nmeth.2400. - PubMed
- Pichlmair A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi:10.1038/nature11289. - PubMed
- Colinge J, Masselot A, Giron M, Dessingy T, Magnin J. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3:1454–1463. doi:10.1002/pmic.200300485. - PubMed
- Bennett KL, et al. Proteomic analysis of human cataract aqueous humour: Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ(R)-labelled specimens. Journal of proteomics. 2011;74:151–166. doi:10.1016/j.jprot.2010.10.002. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous