Activity-Dependent Ubiquitination of GluA1 and GluA2 Regulates AMPA Receptor Intracellular Sorting and Degradation - PubMed (original) (raw)
Activity-Dependent Ubiquitination of GluA1 and GluA2 Regulates AMPA Receptor Intracellular Sorting and Degradation
Jocelyn Widagdo et al. Cell Rep. 2015.
Abstract
AMPA receptors (AMPARs) have recently been shown to undergo post-translational ubiquitination in mammalian neurons. However, the underlying molecular mechanisms are poorly understood and remain controversial. Here, we report that all four AMPAR subunits (GluA1-4) are rapidly ubiquitinated upon brief application of AMPA or bicuculline in cultured neurons. This process is Ca2+ dependent and requires the activity of L-type voltage-gated Ca2+ channels and Ca2+/calmodulin-dependent kinase II. The ubiquitination of all subunits occurs exclusively on AMPARs located on the plasma membrane post-endocytosis. The sites of ubiquitination were mapped to Lys-868 in GluA1 and Lys-870/Lys-882 in GluA2 C-terminals. Mutation of these lysines did not affect basal surface expression or AMPA-induced internalization of GluA1 and GluA2 subunits. Instead, it reduced the intracellular trafficking of AMPARs to the late endosomes and thus protein degradation. These data indicate that ubiquitination is an important regulatory signal for controlling AMPAR function, which may be crucial for synaptic plasticity.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
Figures
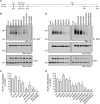
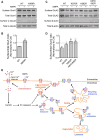
Figure 1. Activity-Dependent Ubiquitination of All AMPAR Subunits Requires Neuronal Depolarization and Ca2+
(A) Cortical neurons were incubated in ACSF in the presence or absence of 100 μM AMPA plus 100 μM D,L-APV and 1 μM TTX for 2 min. The AMPA-treated neurons were immediately incubated in ACSF containing 100 μM D,L-APV and 1 μM TTX for a further 8 min at 37°C. This protocol is referred to as 100 μM AMPA treatment hereafter. Neurons were then lysed in 1% SDS and immunoprecipitated using anti-ubiquitin antibodies (IP: Ub). Eluted proteins were subjected to western blot analysis and probed with anti-ubiquitin and anti-GluA1 antibodies. * denotes non-specific band. (B) Cortical neurons were incubated in ACSF in the absence or presence of glutamate receptor agonists (20 μM NMDA+ 1 μMTTX, 100 μM AMPA + 100 μM D,L-APV and 1 μM TTX, or 100 μM DHPG + 1 μM TTX), 40 μM bicuculline (bic), or 100 μM D,L-APV and 1 μM TTX for 10 min at 37°C. They were then lysed and immunoprecipitated with anti-ubiquitin antibodies and probed with specific antibodies against the GluA1-GluA4 subunits of AMPARs. (C) Surface receptors in cortical neurons were biotinylated at 4°C prior to bicuculline or AMPA stimulation for 10 min at 37°C. Cell lysates were pre-incubated with neutravidin beads prior to immunoprecipitation with anti-ubiquitin antibodies. (D) Cortical neurons were incubated in ACSF containing 2 mM Ca2+ or 0 mM Ca2+ + 50 μM BAPTA-AM + 2 mM EGTA (Ca2+-free) for 20 min before the addition of bicuculline or AMPA for 10 min at 37°C. Cells were lysed and immunoprecipitated with anti-ubiquitin antibodies. (E–H) Cortical neurons were incubated with the indicated inhibitors (vehicle [Veh.], 10 μM NBQX, 10 μM nimodipine[Nimo], 100 μM D,L-APV, and 10 μM Bay 36-7620 + 5 μM MPEP [Bay/MPEP]) for 10 min before adding AMPA(E) or bicuculline (F) for another 10 min at 37°C. (G and H) The effects of these inhibitors on AMPAR ubiquitination were quantified and normalized to vehicle controls (ANOVA; *p < 0.05; **p < 0.01; ***p < 0.001; n = 3–6 per group). Data represent mean ± SEM. (I–L) Cortical neurons were incubated with either DMSO or the CaMKII inhibitor, KN-93(10 μM), for 10 min before adding AMPA (I) or bicuculline (J) for 10 min at 37°C. (K and L) The effects of KN-93 on AMPAR ubiquitination were quantified and normalized to DMSO controls (t test; *p < 0.05; **p < 0.01; n = 3 to 4 per group). Data represent mean ± SEM.
Figure 2. The GluA1 and GluA2 Subunits of AMPARs Are Ubiquitinated on Lysine Residues in Their Carboxy Tails
(A) The amino acid sequence alignment of the GluA1 and GluA2 C termini, with all lysine residues highlighted in black. (B–E) Cortical neurons were electroporated with pH-GluA1 (B) or pH-GluA2 (D) constructs, either wild-type (WT) or mutants in which the lysine residues have been substituted to arginines individually or in combination, prior to plating. At DIV13, the neurons were treated with AMPA for 10min at 37°C, lysed in 1% SDS, and immunoprecipitated using anti-GFP antibodies. Eluted proteins were subjected to western blot analysis and probed with anti-ubiquitin and anti-GluA1 antibodies. The effects of these Lys/Arg mutants on pH-GluA1 (C) and pH-GluA2 (E) ubiquitination were quantified after normalizing against the amount of immunoprecipitated receptor. Data represent mean ± SEM of band intensities normalized to control values of WT neurons (ANOVA; *p < 0.05; **p < 0.01; ***p < 0.001; n = 3 to 4 per group).
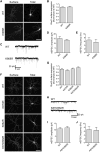
Figure 3. Ubiquitination of GluA1 and GluA2 Does Not Regulate Steady-State Expression of AMPARs
Cultured hippocampal neurons were transfected with pH-GluA1, either WT or the ubiquitin-deficient K868R mutant, at DIV13. At DIV15, surface pH-GluA1 was labeled with rabbit-anti GFP antibody for 30 min at 4°C before fixation. Surface pH-GluA1 was visualized by immunostaining with Alexa Fluor 568 anti-rabbit antibodies, whereas total pH-GluA1 was visualized by the endogenous GFP signal. (A) Representative images of surface and total pH-GluA1 in a neuron from each group. The scale bar represents 50 μm. (B) Quantification of the surface/total pH-GluA1 ratio normalized to the value of WT neurons (n = 31 neurons per group). (C) Representative whole-cell recording sample traces of mEPSC events from each group. (D and E) Quantification of mean mEPSC amplitude (D) and frequency (E) for each population. Data represent mean ± SEM (n = 7 neurons per group). (F–J) Hippocampal neurons transfected with pH-GluA2 ubiquitin-deficient mutants did not display any significant difference in the surface/total receptor ratios (F and G; n = 17–44 neurons per group) or mEPSC amplitude and frequency (H–J; n = 9 to 10 neurons per group). The scale bar represents 50 μm.
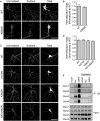
Figure 4. Ubiquitination of GluA1 and GluA2 Is Not Required for the Ligand-Induced Endocytosis of AMPARs
(A) Cultured hippocampal neurons were transfected with pH-GluA1, either WT or ubiquitin-deficient K868R mutant, at DIV13. At DIV15, surface pH-GluA1 was labeled with rabbit anti-GFP antibody for 15 min at 37°C followed by a 10 min incubation with 100 μM AMPA to induce receptor internalization. The remaining surface GFP antibody was stained with Alexa Fluor 568 secondary antibody under non-permeabilizing conditions (surface), and internalized GFP antibody was labeled with Alexa Fluor 647 secondary antibody (internalized). Total pH-GluA1 expression was visualized by the endogenous GFP signal. The scale bar represents 50 μm. (B) The internalization of pH-GluA1 was measured as the ratio of internalized/(internalized + surface) fluorescence (internalization index), normalized to the WT control. Data represent mean ± SEM (n = 11–13 neurons per group). (C and D) Hippocampal neurons transfected with pH-GluA2 ubiquitin-deficient mutants exhibited normal endocytosis following the application of AMPA(C), as shown by the internalization index (D) for each group. Data represent mean ± SEM (n = 27–34 neurons per group). The scale bar in (C) represents 50 μm. (E) Cortical neurons were incubated with an endocytosis inhibitor, dynasore(80 μM), for 10 min before the addition of bicuculline or AMPA for another 10 min at 37°C. Neurons were lysed and immunoprecipitated with anti-ubiquitin antibodies. Eluted proteins were subjected to western blot analysis and probed with specific antibodies against all four AMPAR subunits, GluA1–4.
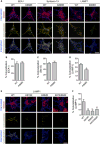
Figure 5. Ubiquitination of GluA1 and GluA2 Regulates the Ligand-Induced Intracellular Sorting of Internalized AMPARs to Late Endosomes
(A) Hippocampal neurons were transiently transfected with pH-GluA1, either WT or the ubiquitin-deficient mutant K868R, at DIV13 for 48 hr. The neurons were then labeled with anti-GFP antibodies, stimulated with AMPA for 2 min, and returned to growth medium for 3 min (early endosomes), 8 min (recycling endosomes), or 28 min (late endosomes) to allow for GluA1 internalization (blue) and sorting into different endosomal compartments (magenta). Simultaneous staining for the early endosome marker, EEA1 (left panels), the recycling endosome marker, syntaxin-13 (middle panels), and the late endosome marker, LAMP1 (right panels), revealed co-localization (yellow) with internalized pH-GluA1 after AMPA application. The scale bar represents 10 μm. (B–D) The extent of association of internalized pH-GluA1 with EEA1- (B), Stx13- (C), and LAMP1-labeled compartments (D) was quantified by image analysis as a percentage of co-localized signals over the amount of total internalized receptors. Data represent mean ± SEM (t test; *p < 0.05; n = 6–14 neurons per group). (E and F) Representative images (E) and quantification (F) of LAMP1-positive endosome co-localization with internalized pH-GluA2, either WT or ubiquitin-deficient mutants (K870R, K882R, and K870/882R), 30 min after AMPA treatment. The scale bar represents 10 μm. Data represent mean ± SEM (ANOVA; *p < 0.05; **p < 0.01; n = 13–16 neurons per group).
Figure 6. Ubiquitination of GluA1 and GluA2 Is Required for Ligand-Induced Degradation of AMPARs
(A–D) Cortical neurons were electroporated with pH-GluA1 (A) and pH-GluA2 (C) constructs, either WT or ubiquitin-deficient mutants as indicated, prior to plating. At DIV13, surface receptors were biotinylated and immediately lysed in RIPA buffer to determine the total amount of biotinylated surface receptors. Sister neuronal cultures were subsequently treated with AMPA for 5 min and returned to growth medium for 90 min at 37°C prior to cell lysis. Neuronal lysates were then incubated with neutravidin beads. Eluted proteins were subjected to western blot analysis and probed with anti-GFP, anti-a-tubulin, and anti-β-actin antibodies. The effects of these mutants on agonist-induced pH-GluA1 (B) and pH-GluA2 (D) degradation were quantified as surface/total receptor ratios and expressed as fractions of remaining receptors after AMPA treatment. Data represent mean ± SEM (t test or ANOVA; *p < 0.05; **p < 0.01; n = 5 to 6 per group). (E) Proposed model for the role of protein ubiquitination in mediating AMPAR endocytic sorting. In this model, activation of AMPARs by the agonist leads to neuronal depolarization and endocytosis of AMPARs into early endosomes. Ligand-induced AMPAR ubiquitination requires Ca2+ influx through L-type voltage-gated Ca2+ channels and CaMKII activity. The latter may activate E3 li-gases such as Nedd4-1, RNF147, or APCCdh1 to ubiquitinate internalized AMPARs, perhaps in the early endosomes. Ubiquitinated receptors are targeted to late endosomes and are subsequently degraded by lysosomes.
Similar articles
- Subunit-Specific Augmentation of AMPA Receptor Ubiquitination by Phorbol Ester.
Widagdo J, Kerk JW, Guntupalli S, Huganir RL, Anggono V. Widagdo J, et al. Cell Mol Neurobiol. 2020 Oct;40(7):1213-1222. doi: 10.1007/s10571-020-00809-2. Epub 2020 Feb 12. Cell Mol Neurobiol. 2020. PMID: 32052226 Free PMC article. - GluA1 subunit ubiquitination mediates amyloid-β-induced loss of surface α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors.
Guntupalli S, Jang SE, Zhu T, Huganir RL, Widagdo J, Anggono V. Guntupalli S, et al. J Biol Chem. 2017 May 19;292(20):8186-8194. doi: 10.1074/jbc.M116.774554. Epub 2017 Apr 4. J Biol Chem. 2017. PMID: 28377502 Free PMC article. - Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway.
Schwarz LA, Hall BJ, Patrick GN. Schwarz LA, et al. J Neurosci. 2010 Dec 8;30(49):16718-29. doi: 10.1523/JNEUROSCI.3686-10.2010. J Neurosci. 2010. PMID: 21148011 Free PMC article. - Activity-dependent ubiquitination of the AMPA receptor subunit GluA2.
Lussier MP, Nasu-Nishimura Y, Roche KW. Lussier MP, et al. J Neurosci. 2011 Feb 23;31(8):3077-81. doi: 10.1523/JNEUROSCI.5944-10.2011. J Neurosci. 2011. PMID: 21414928 Free PMC article. - Regulation of AMPA Receptor Trafficking by Protein Ubiquitination.
Widagdo J, Guntupalli S, Jang SE, Anggono V. Widagdo J, et al. Front Mol Neurosci. 2017 Oct 26;10:347. doi: 10.3389/fnmol.2017.00347. eCollection 2017. Front Mol Neurosci. 2017. PMID: 29123470 Free PMC article. Review.
Cited by
- Pathophysiological Mechanisms of Cognitive Impairment and Neurodegeneration by Toxoplasma gondii Infection.
Ortiz-Guerrero G, Gonzalez-Reyes RE, de-la-Torre A, Medina-Rincón G, Nava-Mesa MO. Ortiz-Guerrero G, et al. Brain Sci. 2020 Jun 12;10(6):369. doi: 10.3390/brainsci10060369. Brain Sci. 2020. PMID: 32545619 Free PMC article. Review. - The AMPA Receptor Code of Synaptic Plasticity.
Diering GH, Huganir RL. Diering GH, et al. Neuron. 2018 Oct 24;100(2):314-329. doi: 10.1016/j.neuron.2018.10.018. Neuron. 2018. PMID: 30359599 Free PMC article. Review. - Loss of fragile X protein FMRP impairs homeostatic synaptic downscaling through tumor suppressor p53 and ubiquitin E3 ligase Nedd4-2.
Lee KY, Jewett KA, Chung HJ, Tsai NP. Lee KY, et al. Hum Mol Genet. 2018 Aug 15;27(16):2805-2816. doi: 10.1093/hmg/ddy189. Hum Mol Genet. 2018. PMID: 29771335 Free PMC article. - Cortactin regulates endo-lysosomal sorting of AMPARs via direct interaction with GluA2 subunit.
Parkinson GT, Chamberlain SEL, Jaafari N, Turvey M, Mellor JR, Hanley JG. Parkinson GT, et al. Sci Rep. 2018 Mar 7;8(1):4155. doi: 10.1038/s41598-018-22542-z. Sci Rep. 2018. PMID: 29515177 Free PMC article. - Methods of measuring presynaptic function with fluorescence probes.
Jang Y, Kim SR, Lee SH. Jang Y, et al. Appl Microsc. 2021 Mar 17;51(1):2. doi: 10.1186/s42649-021-00051-0. Appl Microsc. 2021. PMID: 33730244 Free PMC article. Review.
References
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002;35:107–120. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous