Amyloid precursor protein enhances Nav1.6 sodium channel cell surface expression - PubMed (original) (raw)
Amyloid precursor protein enhances Nav1.6 sodium channel cell surface expression
Chao Liu et al. J Biol Chem. 2015.
Abstract
Amyloid precursor protein (APP) is commonly associated with Alzheimer disease, but its physiological function remains unknown. Nav1.6 is a key determinant of neuronal excitability in vivo. Because mouse models of gain of function and loss of function of APP and Nav1.6 share some similar phenotypes, we hypothesized that APP might be a candidate molecule for sodium channel modulation. Here we report that APP colocalized and interacted with Nav1.6 in mouse cortical neurons. Knocking down APP decreased Nav1.6 sodium channel currents and cell surface expression. APP-induced increases in Nav1.6 cell surface expression were Go protein-dependent, enhanced by a constitutively active Go protein mutant, and blocked by a dominant negative Go protein mutant. APP also regulated JNK activity in a Go protein-dependent manner. JNK inhibition attenuated increases in cell surface expression of Nav1.6 sodium channels induced by overexpression of APP. JNK, in turn, phosphorylated APP. Nav1.6 sodium channel surface expression was increased by T668E and decreased by T668A, mutations of APP695 mimicking and preventing Thr-668 phosphorylation, respectively. Phosphorylation of APP695 at Thr-668 enhanced its interaction with Nav1.6. Therefore, we show that APP enhances Nav1.6 sodium channel cell surface expression through a Go-coupled JNK pathway.
Keywords: Amyloid Precursor Protein (APP); Cell Surface Expression; Duolink; G Protein; Go Protein; JNK; Membrane Protein; Nav1.6 Sodium Channels; Sodium Channel; Trafficking.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
FIGURE 1.
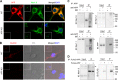
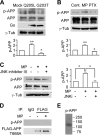
APP interacts with Nav1.6 in vivo and in vitro. A, confocal images of 14 DIV WT and APP KO primary cortical cultures costained with anti-Nav1.6 (Sigma) and anti-APP (Y188, Abcam) antibodies (representative images from 21 cells from three independent cultures). The insets show ×4 lower magnification. Scale bar = 10 μm. B, confocal images showing Duolink fluorescence alone (APP/Nav1.6 interaction, red, left column), differential interference contrast (DIC, center column), and merged with DAPI (right column) in 14 DIV WT and APP KO primary cortical cultures (representative images from 24 cells from three independent cultures). The red dots are positive Duolink signals showing the APP/Nav1.6 interaction. The insets show ×2 lower magnification. Scale bar = 10 μm. C, APP interacts with Nav1.6 in vivo. Adult WT mouse brains were homogenized and solubilized in lysis buffer, and then the soluble extracts were collected by centrifugation at 100,000 × g, immunoprecipitated (IP) using rabbit antibody against the APP C terminus (A8717, Sigma) or normal rabbit IgG control (Cell Signaling Technology), and analyzed by Western blotting using antibodies against Nav1.6, Nav1.2, Nav1.1 (Alomone Laboratories), or APP (MAB348, Millipore) (representative blots from three mice). IB, immunoblot. D, APP interacts with Nav1.6 in vitro. Lysates of HEK293 Nav1.6 cells transfected with pcDNA3-FLAG-APP695 or pcDNA3 collected by centrifugation at 100,000 × g were immunoprecipitated using mouse monoclonal antibody against FLAG and analyzed by Western blotting using antibodies against Nav1.6 (Alomone Laboratories) or FLAG (Sigma) (representative blots from three independent transfections). Input, 5% of total cell lysate; IP, 50% of IP fraction. The arrows indicate the target bands in the multi-band lanes.
FIGURE 2.
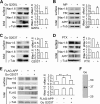
APP enhances Nav1. 6 currents and cell surface expression. A, whole cell recordings of Nav1.6 sodium currents in siRNA control or APP siRNA-transfected HEK293 Nav1.6 cells in which cells were depolarized to a variety of potentials (−80 to +20 mV) from a holding potential of −120 mV. n is indicated in the columns. B–D, total protein and cell surface protein isolated from HEK293 Nav1.6 cells transfected with siRNA control or APP siRNA (B), 14 DIV WT and APP KO primary cortical cultures (C), and mock-transfected and FLAG-APP695 overexpressing Nav1.6 HEK293 cells (D) were blotted using antibodies against Nav1.6, APP, TfR, γ-tubulin (γ_-Tub_), and β3-tubulin (β_3-Tub_). TfR, γ-tubulin, and β3-tubulin served as loading controls for cell surface or total cell lysate fraction, respectively (29). The bar charts show quantitative analysis of relative surface Nav1.6 (sNav1.6) and total Nav1.6 (tNav1.6) expression. The density of the Western blot bands were normalized to the internal loading control and then normalized to control plasmids or treatments. E, representative full immunoblots for the antibodies against TfR, γ-tubulin, and β3-tubulin. Data were analyzed by unpaired Student's t test (A) and paired Student's t tests (B–D). Representative blots from at least three independent experiments were used for statistics. *, p < 0.05; **, p < 0.01. Error bars represent mean ± S.E.
FIGURE 3.
APP promotes Nav1.6 cell surface expression in a Go protein-dependent manner. A–E, total protein and cell surface proteins were isolated from HEK293 Nav1.6 cells transfected with a control plasmid or Go Q205L (A); treated with vehicle or mastoparan (MP, 10 μ
m
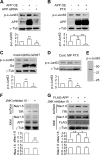
, B); transfected with a control plasmid or Go G203T (C); treated with vehicle or PTX (100 ng/ml, D); transfected with control plasmids, FLAG-APP695 alone, or FLAG-APP695 plus Go 203T (E) were blotted using antibodies against Nav1.6, APP, TfR, and Go. Right panels represent quantitative analysis of relative surface Nav1.6 (sNav1.6) and total Nav1.6 (tNav1.6) expression. γ_-tub_, γ-tubulin. F, representative full immunoblot for the antibody against Go. Representative blots from at least three independent experiments were used for statistics. A–D, paired Student's t test. E, one-way ANOVA followed by post hoc Bonferroni test. *, p < 0.05; **, p < 0.01. Error bars represent mean ± S.E.
FIGURE 4.
APP activates JNK in a Go-protein-dependent manner, and JNK modulates Nav1.6 cell surface expression. A and B, total proteins isolated from HEK293 Nav1.6 cells transfected with APP695 cDNA to induce APP overexpression (APP OE, A), APP siRNA, or control and transfected with APP695 cDNA (APP OE) or control and treated with PTX (100 ng/ml) or vehicle (B) were blotted using antibodies against p-c-Jun Ser-63 (p-c-Jun63), APP, and γ-tubulin (γ_-tub_). C and D, total proteins isolated from HEK293 Nav1.6 cells transfected with Go Q205L or Go G203T or mock-transfected (C) and treated with vehicle control (cont.), mastoparan (MP, 10 μ
m
) or PTX (100 ng/ml) (D) were blotted using antibodies against p-c-Jun63, APP, Go, and γ-tubulin. E, representative full immunoblot for the antibody against p-c-Jun Ser-63. F and G, total proteins and cell surface proteins isolated from HEK293 Nav1.6 cells treated with vehicle or JNK inhibitor III (10 μg/ml, F) or transfected with control plasmids, FLAG-APP695, or FLAG-APP695 and treated with JNK inhibitor III (10 μg/ml, G) were blotted using antibodies against Nav1.6, FLAG, TfR, and γ-tubulin. The bottom panels of A–D represent quantitative analyses of relative p-c-Jun Ser-63 expression by one-way ANOVA followed by post hoc Bonferroni test. The bottom panels of F and G represent quantitative analyses of relative surface Nav1.6 (sNav1.6) and total Nav1.6 (tNav1.6) expression by paired Student's t test or one-way ANOVA followed by post hoc Bonferroni test. Representative blots from at least three independent experiments were used for statistics. *, p < 0.05; **, p < 0.01. Error bars represent mean ± S.E.
FIGURE 5.
Go-protein-dependent Thr-668 phosphorylation of APP695 is mediated by JNK and enhances Nav1.6 sodium channel cell surface expression. A–C, total proteins isolated from HEK293 Nav1.6 cells transfected with Go Q205L or Go G203T or mock-transfected (A); treated with vehicle, mastoparan (MP, 10 μ
m
) or PTX (100 ng/ml) (B); and treated with vehicle or mastoparan (10 μ
m
) or mastoparan (10 μ
m
) plus JNK inhibitor III (JNKi, 10 μg/ml) (C) were blotted using antibodies against p-APP, APP, Go, and γ-tubulin (γ_-tub_). cont., control. D, to show the specificity of rabbit polyclonal antibody against p-APP, lysates of HEK293 Nav1.6 cells transfected with pcDNA3-FLAG-APP695 T668A, treated with vehicle or mastoparan (10 μ
m
), were immunoprecipitated (IP) using normal mouse IgG or mouse monoclonal antibody against FLAG and analyzed by Western blotting using antibody against pAPP. E, representative full immunoblot for the antibody against p-APP. The bar charts in A–C represent quantitative analyses of the relative amount of p-APP by one-way ANOVA followed by post hoc Bonferroni test. Representative blots from at least three independent experiments were used for statistics. *, p < 0.05; **, p < 0.01. Error bars represent mean ± S.E.
FIGURE 6.
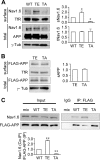
Thr-668 mutants of APP695 influence Nav1.6 surface expression through different interactions with Nav1.6. A, total proteins and cell surface proteins isolated from HEK293 Nav1.6 cells transfected with wild-type APP695 (WT), APP695 T668E (TE, Thr-668 phosphorylation mimic) or APP695 T668A (TA, Thr-668 dephosphorylation mimic) were blotted using antibodies against Nav1.6, APP, TfR, and γ-tubulin (γ_-tub_). The right panel shows a quantitative analysis of relative surface Nav1.6 and total Nav1.6 expression by one-way ANOVA followed by post hoc Bonferroni test. Representative blots from at least three independent experiments were used for statistics. *, p < 0.05. _Error bars_ represent mean ± S.E. _B_, total proteins and cell surface proteins isolated from HEK293 Nav1.6 cells transfected with APP T668E (_TE_) or APP T668A (_TA_) were blotted using antibodies against FLAG, TfR, and γ-tubulin. The _right panel_ shows a quantitative analysis of relative surface APP (_sAPP_). Representative blots from at least three independent experiments were used for statistics (paired Student's _t_ test, _p_ > 0.05). Error bars represent mean ± S.E. C, APP695 T668E interacts with Nav1.6 more strongly than APPT 668A. HEK293 Nav1.6 cells transfected with wild-type APP695 (WT), APP695 T668E, or APP695 T668A were solubilized in lysis buffer. The soluble extracts were collected by centrifugation at 100,000 × g and immunoprecipitated using mouse monoclonal antibody against FLAG (Sigma) or normal mouse IgG (Santa Cruz Biotechnology) added in the same volume mixture of wild-type APP695, APP695 T668E, and APP695 T668A (mix) and analyzed by Western blotting using antibodies against Nav1.6 (Alomone Laboratories) or FLAG (Sigma). The bottom panel shows a quantitative analysis of the relative ratio (normalized to the wild type) of coimmunoprecipitated Nav1.6 versus immunoprecipitated wild-type APP695, APP695 T668E, or APP695 T668A by one-way ANOVA followed by post hoc Bonferroni test. **, p < 0.01. Error bars represent mean ± S.E. Input, 5% of total cell lysate; IP, 50% of immunoprecipitation fraction. Representative blots from at least three independent experiments were used for statistics.
FIGURE 7.
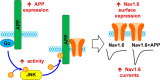
Schematic of the interaction between APP and Nav1.6. Increased expression of APP led to Go protein-dependent activation of JNK. Increased activation of JNK, in turn, led to increased phosphorylation of APP695 at Thr-668. Phosphorylation of APP695 at Thr-668 increased association of APP with Nav1.6, which led to increased cell surface expression of Nav1.6 and, likely, explains greater sodium currents in HEK293 Nav1.6 cells with endogenous APP expression compared with HEK293 Nav1.6 cells in which APP was knocked down.
Similar articles
- Amyloid precursor protein modulates Nav1.6 sodium channel currents through a Go-coupled JNK pathway.
Li S, Wang X, Ma QH, Yang WL, Zhang XG, Dawe GS, Xiao ZC. Li S, et al. Sci Rep. 2016 Dec 23;6:39320. doi: 10.1038/srep39320. Sci Rep. 2016. PMID: 28008944 Free PMC article. - PRRT2 controls neuronal excitability by negatively modulating Na+ channel 1.2/1.6 activity.
Fruscione F, Valente P, Sterlini B, Romei A, Baldassari S, Fadda M, Prestigio C, Giansante G, Sartorelli J, Rossi P, Rubio A, Gambardella A, Nieus T, Broccoli V, Fassio A, Baldelli P, Corradi A, Zara F, Benfenati F. Fruscione F, et al. Brain. 2018 Apr 1;141(4):1000-1016. doi: 10.1093/brain/awy051. Brain. 2018. PMID: 29554219 Free PMC article. - CaMKII enhances voltage-gated sodium channel Nav1.6 activity and neuronal excitability.
Zybura AS, Baucum AJ 2nd, Rush AM, Cummins TR, Hudmon A. Zybura AS, et al. J Biol Chem. 2020 Aug 14;295(33):11845-11865. doi: 10.1074/jbc.RA120.014062. Epub 2020 Jul 1. J Biol Chem. 2020. PMID: 32611770 Free PMC article. - PAT1 inversely regulates the surface Amyloid Precursor Protein level in mouse primary neurons.
Dilsizoglu Senol A, Tagliafierro L, Huguet L, Gorisse-Hussonnois L, Chasseigneaux S, Allinquant B. Dilsizoglu Senol A, et al. BMC Neurosci. 2015 Mar 7;16:10. doi: 10.1186/s12868-015-0152-8. BMC Neurosci. 2015. PMID: 25880931 Free PMC article. - Identification of Amino Acid Residues in Fibroblast Growth Factor 14 (FGF14) Required for Structure-Function Interactions with Voltage-gated Sodium Channel Nav1.6.
Ali SR, Singh AK, Laezza F. Ali SR, et al. J Biol Chem. 2016 May 20;291(21):11268-84. doi: 10.1074/jbc.M115.703868. Epub 2016 Mar 18. J Biol Chem. 2016. PMID: 26994141 Free PMC article.
Cited by
- Analysis of Age-Dependent Alterations in Excitability Properties of CA1 Pyramidal Neurons in an APPPS1 Model of Alzheimer's Disease.
Vitale P, Salgueiro-Pereira AR, Lupascu CA, Willem M, Migliore R, Migliore M, Marie H. Vitale P, et al. Front Aging Neurosci. 2021 Jun 11;13:668948. doi: 10.3389/fnagi.2021.668948. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34177555 Free PMC article. - Alzheimer's-linked axonal changes accompany elevated antidromic action potential failure rate in aged mice.
Russo ML, Ayala G, Neal D, Rogalsky AE, Ahmad S, Musial TF, Pearlman M, Bean LA, Farooqi AK, Ahmed A, Castaneda A, Patel A, Parduhn Z, Haddad LG, Gabriel A, Disterhoft JF, Nicholson DA. Russo ML, et al. Brain Res. 2024 Oct 15;1841:149083. doi: 10.1016/j.brainres.2024.149083. Epub 2024 Jun 10. Brain Res. 2024. PMID: 38866308 - Voltage-Gated Sodium Channel Dysfunctions in Neurological Disorders.
Barbieri R, Nizzari M, Zanardi I, Pusch M, Gavazzo P. Barbieri R, et al. Life (Basel). 2023 May 16;13(5):1191. doi: 10.3390/life13051191. Life (Basel). 2023. PMID: 37240836 Free PMC article. Review. - Sodium channel subtypes are differentially localized to pre- and post-synaptic sites in rat hippocampus.
Johnson KW, Herold KF, Milner TA, Hemmings HC Jr, Platholi J. Johnson KW, et al. J Comp Neurol. 2017 Nov 1;525(16):3563-3578. doi: 10.1002/cne.24291. Epub 2017 Aug 11. J Comp Neurol. 2017. PMID: 28758202 Free PMC article. - Voltage-Gated Na+ Channels in Alzheimer's Disease: Physiological Roles and Therapeutic Potential.
Baumgartner TJ, Haghighijoo Z, Goode NA, Dvorak NM, Arman P, Laezza F. Baumgartner TJ, et al. Life (Basel). 2023 Jul 29;13(8):1655. doi: 10.3390/life13081655. Life (Basel). 2023. PMID: 37629512 Free PMC article. Review.
References
- Turner P. R., O'Connor K., Tate W. P., Abraham W. C. (2003) Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog. Neurobiol. 70, 1–32 - PubMed
- Duce J. A., Tsatsanis A., Cater M. A., James S. A., Robb E., Wikhe K., Leong S. L., Perez K., Johanssen T., Greenough M. A., Cho H. H., Galatis D., Moir R. D., Masters C. L., McLean C., Tanzi R. E., Cappai R., Barnham K. J., Ciccotosto G. D., Rogers J. T., Bush A. I. (2010) Iron-export ferroxidase activity of β-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell 142, 857–867 - PMC - PubMed
- Zheng H., Jiang M., Trumbauer M. E., Sirinathsinghji D. J., Hopkins R., Smith D. W., Heavens R. P., Dawson G. R., Boyce S., Conner M. W., Stevens K. A., Slunt H. H., Sisoda S. S., Chen H. Y., Van der Ploeg L. H. (1995) β-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81, 525–531 - PubMed
- Busche M. A., Eichhoff G., Adelsberger H., Abramowski D., Wiederhold K. H., Haass C., Staufenbiel M., Konnerth A., Garaschuk O. (2008) Clusters of hyperactive neurons near amyloid plaques in a mouse model of Alzheimer's disease. Science 321, 1686–1689 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials