Antagonizing pathways leading to differential dynamics in colon carcinogenesis in Shugoshin1 (Sgo1)-haploinsufficient chromosome instability model - PubMed (original) (raw)
Antagonizing pathways leading to differential dynamics in colon carcinogenesis in Shugoshin1 (Sgo1)-haploinsufficient chromosome instability model
Chinthalapally V Rao et al. Mol Carcinog. 2016 May.
Abstract
Colon cancer is the second most lethal cancer. It is predicted to claim 50,310 lives in 2014. Chromosome Instability (CIN) is observed in 80-90% of colon cancers, and is thought to contribute to colon cancer progression and recurrence. However, there are no animal models of CIN that have been validated for studies of colon cancer development or drug testing. In this study, we sought to validate a mitotic error-induced CIN model mouse, the Shugoshin1 (Sgo1) haploinsufficient mouse, as a colon cancer study model. Wild-type and Sgo1(-/+) mice were treated with the colonic carcinogen, azoxymethane (AOM). We tracked colon tumor development 12, 24, and 36 wk after treatment to assess progression of colon tumorigenesis. Initially, more precancerous lesions, Aberrant Crypt Foci (ACF), developed in Sgo1(-/+) mice. However, the ACF did not develop straightforwardly into larger tumors. At the 36-wk endpoint, the number of gross tumors in Sgo1(-/+) mice was no different from that in wild-type controls. However, Copy Number Variation (CNV) analysis indicated that fully developed colon tumor in Sgo1(-/+) mice carried 13.75 times more CNV. Immunohistological analyses indicated that Sgo1(-/+) mice differentially expressed IL-6, Bcl2, and p16(INK4A) . We propose that formation of ACF in Sgo1(-/+) mice is facilitated by the IL6-STAT3-SOCS3 oncogenic pathway and by the Bcl2-anti-apoptotic pathway, yet further development of the ACF to tumors is inhibited by the p16(INK4A) tumor suppressor pathway. Manipulating these pathways would be beneficial for inhibiting development of colon cancer with CIN.
Keywords: Azoxymethane (AOM); Chromosome Instability (CIN); Colon cancer; Sgo1 (Shugoshin 1); mice.
© 2015 Wiley Periodicals, Inc.
Figures
Fig 1. Experimental scheme
(A) Samples were collected at three endpoints (12 weeks, 24 weeks, and 36 weeks post AOM treatments). For gross tumor formation, see Table 1. Only limited and comparable numbers of colon tumors developed at the endpoints. No loss of animals was observed during the entire experiment. (B) Average body weight at each endpoint showed no statistically significant difference. _N_=15 for wild-type (green) and for Sgo1 (red).
Fig2. Different dynamics in the development of colonic ACFs and tumors (microadenomas) in wild-type and Sgo1 mice
(A) At endpoint 1 (12 weeks), the total ACF number per animal was significantly higher in Sgo1 mice, indicating rapid development of lesions in Sgo1. (B) At endpoint 2 (24 weeks), wild-type mice kept developing ACF, yet further development of ACF in Sgo1 mice was not observed. The total ACF number per animal was significantly less in Sgo1 mice. Numbers of microscopic tumors (microadenomas) showed no difference (ns: non-significant). (C) At endpoint 3 (36 weeks), most ACF regressed in both strains.
Fig2. Different dynamics in the development of colonic ACFs and tumors (microadenomas) in wild-type and Sgo1 mice
(A) At endpoint 1 (12 weeks), the total ACF number per animal was significantly higher in Sgo1 mice, indicating rapid development of lesions in Sgo1. (B) At endpoint 2 (24 weeks), wild-type mice kept developing ACF, yet further development of ACF in Sgo1 mice was not observed. The total ACF number per animal was significantly less in Sgo1 mice. Numbers of microscopic tumors (microadenomas) showed no difference (ns: non-significant). (C) At endpoint 3 (36 weeks), most ACF regressed in both strains.
Fig2. Different dynamics in the development of colonic ACFs and tumors (microadenomas) in wild-type and Sgo1 mice
(A) At endpoint 1 (12 weeks), the total ACF number per animal was significantly higher in Sgo1 mice, indicating rapid development of lesions in Sgo1. (B) At endpoint 2 (24 weeks), wild-type mice kept developing ACF, yet further development of ACF in Sgo1 mice was not observed. The total ACF number per animal was significantly less in Sgo1 mice. Numbers of microscopic tumors (microadenomas) showed no difference (ns: non-significant). (C) At endpoint 3 (36 weeks), most ACF regressed in both strains.
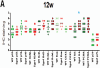
Fig3. Differentially expressed proteins in Sgo1 colon
Paraffin-embedded colon sections were stained with the indicated antibody. Samples from wild-type mice are indicated in green, Sgo1 mice in red. IHC signals were graded in 7 grades (See Materials and Methods) for indicated markers. Asterisk (blue) indicates more than 1 grade scale difference in expression. (A) 12 weeks endpoint. IL-6 was expressed notably higher in Sgo1 mice. (B) 24 weeks endpoint. p16, Bcl2, and IL-6 were notably higher in Sgo1 mice. (C) 36 weeks endpoint. Bcl2, BclxL, Bax, IL-6, and p21 were notably higher in Sgo1 mice.
Fig3. Differentially expressed proteins in Sgo1 colon
Paraffin-embedded colon sections were stained with the indicated antibody. Samples from wild-type mice are indicated in green, Sgo1 mice in red. IHC signals were graded in 7 grades (See Materials and Methods) for indicated markers. Asterisk (blue) indicates more than 1 grade scale difference in expression. (A) 12 weeks endpoint. IL-6 was expressed notably higher in Sgo1 mice. (B) 24 weeks endpoint. p16, Bcl2, and IL-6 were notably higher in Sgo1 mice. (C) 36 weeks endpoint. Bcl2, BclxL, Bax, IL-6, and p21 were notably higher in Sgo1 mice.
Fig3. Differentially expressed proteins in Sgo1 colon
Paraffin-embedded colon sections were stained with the indicated antibody. Samples from wild-type mice are indicated in green, Sgo1 mice in red. IHC signals were graded in 7 grades (See Materials and Methods) for indicated markers. Asterisk (blue) indicates more than 1 grade scale difference in expression. (A) 12 weeks endpoint. IL-6 was expressed notably higher in Sgo1 mice. (B) 24 weeks endpoint. p16, Bcl2, and IL-6 were notably higher in Sgo1 mice. (C) 36 weeks endpoint. Bcl2, BclxL, Bax, IL-6, and p21 were notably higher in Sgo1 mice.
Fig 4. Different patterns of IL-6, Bcl2, and p16INK4A protein localization in wild-type and Sgo1 mice
(A) IL-6 expression was generally low in normal-looking mucosa in Sgo1−/+ and wild-type controls. Lesions in wild-type mice sporadically showed IL-6-positive cells. The signals were prominent in lesions in Sgo1−/+. (B) Bcl2 expression was higher in normal-looking parts of colons in Sgo1−/+ mice than in wild-type mice. As lesions developed, Bcl2 expression increased in both Sgo1−/+ and wild-type mice. (C) p16INK4A expression was low in wild-type mice, but higher in Sgo1−/+ mice, in both normal-looking colons and in lesions. [(A)-(C): all ACF/lesions were confirmed by a histopathologist]. (D) Immunoblots for the markers; IL6/STAT3/SOCS3 pathway (IL6, IL6 receptor (IL6R), phosphor-STAT3, SOCS3), apoptosis pathway (Bcl2, Bax), tumor suppressor pathway (p53, p21, p16INK4A). Samples from Sgo1 (lanes 7-12) tend to show an increase in marker protein expression compared with samples from wild type (lanes 1-6), in agreement with IHC results. (E) The differences in protein amount may not be due to transcriptional upregulation. NGS readouts (left bar: wild type; right bar: Sgo1) did not show significant up-regulation in mRNA level for indicated markers. Beta-actin and pgk1 (Phosphoglycerate kinase 1) are commonly used for qPCR controls. The readouts are shown as NGS quantification controls. ns: non-significant.
Fig 4. Different patterns of IL-6, Bcl2, and p16INK4A protein localization in wild-type and Sgo1 mice
(A) IL-6 expression was generally low in normal-looking mucosa in Sgo1−/+ and wild-type controls. Lesions in wild-type mice sporadically showed IL-6-positive cells. The signals were prominent in lesions in Sgo1−/+. (B) Bcl2 expression was higher in normal-looking parts of colons in Sgo1−/+ mice than in wild-type mice. As lesions developed, Bcl2 expression increased in both Sgo1−/+ and wild-type mice. (C) p16INK4A expression was low in wild-type mice, but higher in Sgo1−/+ mice, in both normal-looking colons and in lesions. [(A)-(C): all ACF/lesions were confirmed by a histopathologist]. (D) Immunoblots for the markers; IL6/STAT3/SOCS3 pathway (IL6, IL6 receptor (IL6R), phosphor-STAT3, SOCS3), apoptosis pathway (Bcl2, Bax), tumor suppressor pathway (p53, p21, p16INK4A). Samples from Sgo1 (lanes 7-12) tend to show an increase in marker protein expression compared with samples from wild type (lanes 1-6), in agreement with IHC results. (E) The differences in protein amount may not be due to transcriptional upregulation. NGS readouts (left bar: wild type; right bar: Sgo1) did not show significant up-regulation in mRNA level for indicated markers. Beta-actin and pgk1 (Phosphoglycerate kinase 1) are commonly used for qPCR controls. The readouts are shown as NGS quantification controls. ns: non-significant.
Fig 4. Different patterns of IL-6, Bcl2, and p16INK4A protein localization in wild-type and Sgo1 mice
(A) IL-6 expression was generally low in normal-looking mucosa in Sgo1−/+ and wild-type controls. Lesions in wild-type mice sporadically showed IL-6-positive cells. The signals were prominent in lesions in Sgo1−/+. (B) Bcl2 expression was higher in normal-looking parts of colons in Sgo1−/+ mice than in wild-type mice. As lesions developed, Bcl2 expression increased in both Sgo1−/+ and wild-type mice. (C) p16INK4A expression was low in wild-type mice, but higher in Sgo1−/+ mice, in both normal-looking colons and in lesions. [(A)-(C): all ACF/lesions were confirmed by a histopathologist]. (D) Immunoblots for the markers; IL6/STAT3/SOCS3 pathway (IL6, IL6 receptor (IL6R), phosphor-STAT3, SOCS3), apoptosis pathway (Bcl2, Bax), tumor suppressor pathway (p53, p21, p16INK4A). Samples from Sgo1 (lanes 7-12) tend to show an increase in marker protein expression compared with samples from wild type (lanes 1-6), in agreement with IHC results. (E) The differences in protein amount may not be due to transcriptional upregulation. NGS readouts (left bar: wild type; right bar: Sgo1) did not show significant up-regulation in mRNA level for indicated markers. Beta-actin and pgk1 (Phosphoglycerate kinase 1) are commonly used for qPCR controls. The readouts are shown as NGS quantification controls. ns: non-significant.
Fig 4. Different patterns of IL-6, Bcl2, and p16INK4A protein localization in wild-type and Sgo1 mice
(A) IL-6 expression was generally low in normal-looking mucosa in Sgo1−/+ and wild-type controls. Lesions in wild-type mice sporadically showed IL-6-positive cells. The signals were prominent in lesions in Sgo1−/+. (B) Bcl2 expression was higher in normal-looking parts of colons in Sgo1−/+ mice than in wild-type mice. As lesions developed, Bcl2 expression increased in both Sgo1−/+ and wild-type mice. (C) p16INK4A expression was low in wild-type mice, but higher in Sgo1−/+ mice, in both normal-looking colons and in lesions. [(A)-(C): all ACF/lesions were confirmed by a histopathologist]. (D) Immunoblots for the markers; IL6/STAT3/SOCS3 pathway (IL6, IL6 receptor (IL6R), phosphor-STAT3, SOCS3), apoptosis pathway (Bcl2, Bax), tumor suppressor pathway (p53, p21, p16INK4A). Samples from Sgo1 (lanes 7-12) tend to show an increase in marker protein expression compared with samples from wild type (lanes 1-6), in agreement with IHC results. (E) The differences in protein amount may not be due to transcriptional upregulation. NGS readouts (left bar: wild type; right bar: Sgo1) did not show significant up-regulation in mRNA level for indicated markers. Beta-actin and pgk1 (Phosphoglycerate kinase 1) are commonly used for qPCR controls. The readouts are shown as NGS quantification controls. ns: non-significant.
Fig 4. Different patterns of IL-6, Bcl2, and p16INK4A protein localization in wild-type and Sgo1 mice
(A) IL-6 expression was generally low in normal-looking mucosa in Sgo1−/+ and wild-type controls. Lesions in wild-type mice sporadically showed IL-6-positive cells. The signals were prominent in lesions in Sgo1−/+. (B) Bcl2 expression was higher in normal-looking parts of colons in Sgo1−/+ mice than in wild-type mice. As lesions developed, Bcl2 expression increased in both Sgo1−/+ and wild-type mice. (C) p16INK4A expression was low in wild-type mice, but higher in Sgo1−/+ mice, in both normal-looking colons and in lesions. [(A)-(C): all ACF/lesions were confirmed by a histopathologist]. (D) Immunoblots for the markers; IL6/STAT3/SOCS3 pathway (IL6, IL6 receptor (IL6R), phosphor-STAT3, SOCS3), apoptosis pathway (Bcl2, Bax), tumor suppressor pathway (p53, p21, p16INK4A). Samples from Sgo1 (lanes 7-12) tend to show an increase in marker protein expression compared with samples from wild type (lanes 1-6), in agreement with IHC results. (E) The differences in protein amount may not be due to transcriptional upregulation. NGS readouts (left bar: wild type; right bar: Sgo1) did not show significant up-regulation in mRNA level for indicated markers. Beta-actin and pgk1 (Phosphoglycerate kinase 1) are commonly used for qPCR controls. The readouts are shown as NGS quantification controls. ns: non-significant.
Fig 5. Tumors in Sgo1 mice had higher CNV than tumors in wild-type mice
(A) Normal-looking mucosal tissues vs tumors [wild-type]. 12 CNV were identified in the tumor. Red: amplification. Green: loss. (B) Normal-looking mucosal tissues vs tumors [Sgo1]. 165 CNV were identified in the tumor. (C) Wild-type tumors vs Sgo1 tumors. 40 CNV were identified. The CNV analysis was array/hybridization-based, and would detect regional loss/gain in chromosomes as well as whole chromosome loss/gain if happened in a majority of the tumor cells. However, it should be noted that polyploidization (e.g. tertaploidization) may not be detected well in this method. Since we used female mice for the experiments, no Y chromosome probes appeared positive in this analysis.
Fig 5. Tumors in Sgo1 mice had higher CNV than tumors in wild-type mice
(A) Normal-looking mucosal tissues vs tumors [wild-type]. 12 CNV were identified in the tumor. Red: amplification. Green: loss. (B) Normal-looking mucosal tissues vs tumors [Sgo1]. 165 CNV were identified in the tumor. (C) Wild-type tumors vs Sgo1 tumors. 40 CNV were identified. The CNV analysis was array/hybridization-based, and would detect regional loss/gain in chromosomes as well as whole chromosome loss/gain if happened in a majority of the tumor cells. However, it should be noted that polyploidization (e.g. tertaploidization) may not be detected well in this method. Since we used female mice for the experiments, no Y chromosome probes appeared positive in this analysis.
Fig 5. Tumors in Sgo1 mice had higher CNV than tumors in wild-type mice
(A) Normal-looking mucosal tissues vs tumors [wild-type]. 12 CNV were identified in the tumor. Red: amplification. Green: loss. (B) Normal-looking mucosal tissues vs tumors [Sgo1]. 165 CNV were identified in the tumor. (C) Wild-type tumors vs Sgo1 tumors. 40 CNV were identified. The CNV analysis was array/hybridization-based, and would detect regional loss/gain in chromosomes as well as whole chromosome loss/gain if happened in a majority of the tumor cells. However, it should be noted that polyploidization (e.g. tertaploidization) may not be detected well in this method. Since we used female mice for the experiments, no Y chromosome probes appeared positive in this analysis.
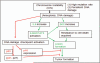
Fig 6. Model for differential dynamics in lesion/tumor development with Sgo1 defects
Current hypothetical model for how Sgo1 defect-mediated CIN leads to differential dynamics in colon lesion and tumor development. CIN leads to a high mutation rate and DNA damage, resulting in aneuploidy and/or DNA damage (16, 42, 43). As a result, the DNA damage checkpoint is activated and senescence is induced. The senescence serves to inhibit lesion/tumor growth. On the other hand, DNA damage leads to activation of IL-6 pathway. In addition, the Bcl2/Bax/BclxL-mediated cell death pathway is affected and a resistance to cell death is acquired. These two events drive initial lesion and tumor formation. The differential dynamics in the high CIN condition are a result of these antagonizing pathways. Formation process of tumors with high CIN may be modulated by interventions in these pathways.
Similar articles
- Systemic Chromosome Instability Resulted in Colonic Transcriptomic Changes in Metabolic, Proliferation, and Stem Cell Regulators in Sgo1-/+ Mice.
Rao CV, Sanghera S, Zhang Y, Biddick L, Reddy A, Lightfoot S, Janakiram NB, Mohammed A, Dai W, Yamada HY. Rao CV, et al. Cancer Res. 2016 Feb 1;76(3):630-42. doi: 10.1158/0008-5472.CAN-15-0940. Cancer Res. 2016. PMID: 26833665 Free PMC article. - Haploinsufficiency of SGO1 results in deregulated centrosome dynamics, enhanced chromosomal instability and colon tumorigenesis.
Yamada HY, Yao Y, Wang X, Zhang Y, Huang Y, Dai W, Rao CV. Yamada HY, et al. Cell Cycle. 2012 Feb 1;11(3):479-88. doi: 10.4161/cc.11.3.18994. Epub 2012 Feb 1. Cell Cycle. 2012. PMID: 22262168 Free PMC article. - Tumor-promoting/progressing role of additional chromosome instability in hepatic carcinogenesis in Sgo1 (Shugoshin 1) haploinsufficient mice.
Yamada HY, Zhang Y, Reddy A, Mohammed A, Lightfoot S, Dai W, Rao CV. Yamada HY, et al. Carcinogenesis. 2015 Apr;36(4):429-40. doi: 10.1093/carcin/bgv011. Epub 2015 Mar 4. Carcinogenesis. 2015. PMID: 25740822 Free PMC article. - Emerging links among Chromosome Instability (CIN), cancer, and aging.
Rao CV, Asch AS, Yamada HY. Rao CV, et al. Mol Carcinog. 2017 Mar;56(3):791-803. doi: 10.1002/mc.22539. Epub 2016 Aug 30. Mol Carcinog. 2017. PMID: 27533343 Review. - Inhibition of aberrant crypt foci by chemopreventive agents.
Olivo S, Wargovich MJ. Olivo S, et al. In Vivo. 1998 Mar-Apr;12(2):159-66. In Vivo. 1998. PMID: 9627797 Review.
Cited by
- Shugoshin 1 expression in various cancers: a potential target for therapy.
Ankathatti Narayanaswamy I, Kattepur AK, Raju K, Perumal V, Ramalingam R, Raavi V. Ankathatti Narayanaswamy I, et al. Clin Transl Oncol. 2024 Oct 30. doi: 10.1007/s12094-024-03749-1. Online ahead of print. Clin Transl Oncol. 2024. PMID: 39476301 Review. - The expanding phenotypes of cohesinopathies: one ring to rule them all!
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. Piché J, et al. Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review. - Critical role of mitosis in spontaneous late-onset Alzheimer's disease; from a Shugoshin 1 cohesinopathy mouse model.
Rao CV, Farooqui M, Asch AS, Yamada HY. Rao CV, et al. Cell Cycle. 2018;17(19-20):2321-2334. doi: 10.1080/15384101.2018.1515554. Epub 2018 Sep 20. Cell Cycle. 2018. PMID: 30231670 Free PMC article. Review. - Systemic Chromosome Instability Resulted in Colonic Transcriptomic Changes in Metabolic, Proliferation, and Stem Cell Regulators in Sgo1-/+ Mice.
Rao CV, Sanghera S, Zhang Y, Biddick L, Reddy A, Lightfoot S, Janakiram NB, Mohammed A, Dai W, Yamada HY. Rao CV, et al. Cancer Res. 2016 Feb 1;76(3):630-42. doi: 10.1158/0008-5472.CAN-15-0940. Cancer Res. 2016. PMID: 26833665 Free PMC article. - Systemic chromosome instability in Shugoshin-1 mice resulted in compromised glutathione pathway, activation of Wnt signaling and defects in immune system in the lung.
Yamada HY, Kumar G, Zhang Y, Rubin E, Lightfoot S, Dai W, Rao CV. Yamada HY, et al. Oncogenesis. 2016 Aug 15;5(8):e256. doi: 10.1038/oncsis.2016.56. Oncogenesis. 2016. PMID: 27526110 Free PMC article.
References
- American Cancer Society (ACS) statistics for colon cancer 2014 http://www.cancer.org/cancer/colonandrectumcancer/detailedguide/colorect....
- Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21(20):3253–7. - PubMed
- Chandhok NS, Pellman D. A little CIN may cost a lot: revisiting aneuploidy and cancer. Curr Opin Genet Dev. 2009;19(1):74–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 CA094962/CA/NCI NIH HHS/United States
- R01 CA090658/CA/NCI NIH HHS/United States
- R03 CA162538/CA/NCI NIH HHS/United States
- R01 ES020591/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous