Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase - PubMed (original) (raw)
Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase
Christopher L Holley et al. J Biol Chem. 2015.
Abstract
Small nucleolar RNAs (snoRNAs) guide nucleotide modifications of cellular RNAs in the nucleus. We previously showed that box C/D snoRNAs from the Rpl13a locus are unexpected mediators of physiologic oxidative stress, independent of their predicted ribosomal RNA modifications. Here we demonstrate that oxidative stress induced by doxorubicin causes rapid cytoplasmic accumulation of the Rpl13a snoRNAs through a mechanism that requires superoxide and a nuclear splice variant of NADPH oxidase. RNA-sequencing analysis reveals that box C/D snoRNAs as a class are present in the cytoplasm, where their levels are dynamically regulated by NADPH oxidase. These findings suggest that snoRNAs may orchestrate the response to environmental stress through molecular interactions outside of the nucleus.
Keywords: Doxorubicin; Intracellular Trafficking; Molecular Cell Biology; NADPH Oxidase; Reactive Oxygen Species (ROS); Small Nucleolar RNA (snoRNA); Subcellular Fractionation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
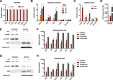
FIGURE 1.
Dox stimulates rapid cytosolic accumulation of Rpl13a snoRNAs. A, H9c2 cells were treated with 20 μ
m
dox for 1 h, and total cellular RNA was assayed by qPCR. Relative quantification is shown, with normalization to the control transcript Rplp0. Error bars show 95% confidence interval for n = 3 experiments. ctrl, control. B, H9c2 cells were treated with 20 μ
m
dox for the times shown. Cytosol was isolated by selective permeabilization of the plasma membrane with digitonin buffer (detergent extraction) and quantified as in A. *, p < 0.05 versus control. C, cells were treated with dox for 1 h as in B or untreated (ctrl), and cytosol was isolated by detergent-free hypotonic lysis. Cytosolic snoRNA was quantified as in B. D, cytotoxicity was assayed by lactate dehydrogenase (LDH) release into the media after 1 h of treatment with 20 μ
m
dox. Cell lysis to release total cellular lactate dehydrogenase served as maximal cytotoxicity (max). The graph shows mean absorbance at 492 nm + S.E. for n = 3 experiments. *, p < 0.05 versus control. E–H, cytosolic and nuclear extracts were prepared from control and 1-h dox-treated cells by detergent extraction (E and F) and by hypotonic lysis (G and H). Fractions were analyzed by Western blot for protein markers of cytoplasm (Hsp90, α-tubulin), nucleus (histone H3), and nucleolus (NPM) (E and G). qPCR quantification of nuclear ncRNAs (7SK and U6) and the Rpl13a snoRNAs shows the relative amounts of RNA from cytosolic and nuclear fractions (F and H, note the log scale; *, p < 0.05 versus control for cytoplasm; #, p < 0.05 versus control for nuclear).
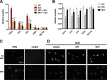
FIGURE 2.
Superoxide production is necessary for cytosolic accumulation of Rpl13a snoRNAs in response to dox. A, H9c2 cells were treated for 1 h with either vehicle alone (ctrl) or combinations of 20 μ
m
dox, 500 n
m
DPI, and 200 μ
m
MnT as shown. Cytosolic RNA was isolated by detergent extraction and quantified relative to Rplp0 by qPCR. Error bars show 95% confidence interval for n = 3 experiments. B, H9c2 cells were treated with DPI or MnT alone. Cytosol was isolated and analyzed for cytosolic snoRNAs as in A. C and D, H9c2 cells were analyzed by immunofluorescence microscopy for detection of NPM under control and 1 h dox-treated conditions and also in the presence of DPI or MnT as in A. Micrographs show representative fields. Scale bar, 200 μm. *, p < 0.05 versus control; #, p < 0.05 versus dox alone.
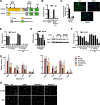
FIGURE 3.
NAPDH oxidase mediates cytosolic accumulation of Rpl13a snoRNAs in response to dox. A, the exonic structures of Nox4A and alternatively-spliced nuclear form Nox4D are depicted, with localization of siRNAs that target Nox4A only (exons 3 and 5) or both Nox4A and D (exons 12–13 and 14). B–G, H9c2 cells were transfected for 24 h with siRNA targeting transcripts as shown: control, p22, Nox4A/D, or Nox4A. Cells were then treated with 20 μ
m
dox for 1 h where indicated. RNAs were quantified by qPCR and are reported as mean values relative to Rplp0 for n = 3 experiments. Error bars show 95% confidence interval. Total RNA was assessed for target knockdown with and without dox treatment (B, D, and F). Cells were analyzed by immunofluorescence microscopy to confirm knockdown of p22 protein (C). Fluorescence intensity under identical imaging conditions was quantified, and the graph shows average intensity per cell (>50 cells per condition). Representative micrographs are shown; green = p22; blue = Hoechst. Cell lysates were analyzed by Western blot to confirm knockdown of the 67kD Nox4A isoform (E). Rat Nox4D was not detected by any commercially available antibodies. The graph shows densitometry quantification of n = 3 independent experiments with normalization to Hsp90. A representative blot is shown. Cytosolic RNA was isolated by detergent extraction, and Rpl13a snoRNAs were quantified by qPCR (G). H, cells were treated as above and analyzed by immunofluorescence microscopy for localization of NPM. Micrographs show representative fields. Scale bar, 200 μm. *, p < 0.05 versus control; #, p < 0.05 versus control/dox; &, p < 0.05 versus Nox4A/dox.
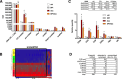
FIGURE 4.
snoRNAs as a class are dynamically regulated in the cytoplasm. H9c2 cells were treated with dox (20 μ
m
) and/or DPI (500 n
m
) for 1 h. A, normalized RNA-seq read counts for annotated cytoplasmic small ncRNA classes. Annotated reads were normalized as reads per million raw reads (RpM) and shown as the mean ± S.E. for n = 3 experiments. ctrl, control. B, unsupervised hierarchical clustering of 132 annotated ncRNA used reads from A to generate a heat map showing relative changes in cytoplasmic small ncRNA levels with the treatments. The black box highlights clustering of snoRNAs and scaRNAs. Reads mapping to miRNAs were omitted for clarity. C, RNA-seq expression data for cytosolic U6, 7SK, and Rpl13a snoRNAs in response to treatment with DPI, dox, or DPI/dox. Read data are shown as reads/million aligned reads to demonstrate the relative abundance of different Rpl13a snoRNA species (upper). These values were then further normalized to the control sample and graphed to show relative expression by treatment (lower; mean values + S.E.; n = 3 samples). D, 105 small ncRNA genes in the cytoplasm were differentially expressed after dox treatment. The number of genes annotated for the rat genome (Ensembl ncRNA), number of genes found experimentally by cytoplasmic RNA-seq (detected in cytoplasm), and number of genes detected in the cytoplasm and up-regulated by dox (versus control) are shown by ncRNA subclass. *, p < 0.05 for treatment versus control; #, p < 0.05 for DPI/dox versus dox alone.
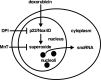
FIGURE 5.
Proposed model showing regulation of cytosolic snoRNA abundance by Nox4D and superoxide in response to doxorubicin.
Similar articles
- Nuclear export factor 3 regulates localization of small nucleolar RNAs.
Li MW, Sletten AC, Lee J, Pyles KD, Matkovich SJ, Ory DS, Schaffer JE. Li MW, et al. J Biol Chem. 2017 Dec 8;292(49):20228-20239. doi: 10.1074/jbc.M117.818146. Epub 2017 Oct 11. J Biol Chem. 2017. PMID: 29021253 Free PMC article. - Small nucleolar RNA of silkworm can translocate from the nucleolus to the cytoplasm under abiotic stress.
Huo CY, Chang ML, Cheng H, Ma TT, Fu Y, Wang Y, Wang YY, Kan YC, Li DD. Huo CY, et al. Cell Biol Int. 2021 May;45(5):1091-1097. doi: 10.1002/cbin.11555. Epub 2021 Feb 2. Cell Biol Int. 2021. PMID: 33501699 - Long-range function of secreted small nucleolar RNAs that direct 2'-_O_-methylation.
Rimer JM, Lee J, Holley CL, Crowder RJ, Chen DL, Hanson PI, Ory DS, Schaffer JE. Rimer JM, et al. J Biol Chem. 2018 Aug 24;293(34):13284-13296. doi: 10.1074/jbc.RA118.003410. Epub 2018 Jul 6. J Biol Chem. 2018. PMID: 29980600 Free PMC article. - Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin.
Terns MP, Terns RM. Terns MP, et al. Gene Expr. 2002;10(1-2):17-39. Gene Expr. 2002. PMID: 11868985 Free PMC article. Review. - The regulatory roles of small nucleolar RNAs within their host locus.
Fafard-Couture É, Labialle S, Scott MS. Fafard-Couture É, et al. RNA Biol. 2024 Jan;21(1):1-11. doi: 10.1080/15476286.2024.2342685. Epub 2024 Apr 16. RNA Biol. 2024. PMID: 38626213 Free PMC article. Review.
Cited by
- Annotation of snoRNA abundance across human tissues reveals complex snoRNA-host gene relationships.
Fafard-Couture É, Bergeron D, Couture S, Abou-Elela S, Scott MS. Fafard-Couture É, et al. Genome Biol. 2021 Jun 4;22(1):172. doi: 10.1186/s13059-021-02391-2. Genome Biol. 2021. PMID: 34088344 Free PMC article. - The Arabidopsis 2'-O-Ribose-Methylation and Pseudouridylation Landscape of rRNA in Comparison to Human and Yeast.
Streit D, Schleiff E. Streit D, et al. Front Plant Sci. 2021 Jul 26;12:684626. doi: 10.3389/fpls.2021.684626. eCollection 2021. Front Plant Sci. 2021. PMID: 34381476 Free PMC article. Review. - Rpl13a snoRNAs-regulated NADPH oxidase 1-dependent ROS generation: A novel RBC pathway mediating complement C3a deposition and triggering thrombosis in aging and venous blood clotting disorders.
Chauhan W, Ferdowsi S, Sudharshan SJ, Zennadi R. Chauhan W, et al. Free Radic Biol Med. 2025 Mar 16;230:138-150. doi: 10.1016/j.freeradbiomed.2025.02.008. Epub 2025 Feb 10. Free Radic Biol Med. 2025. PMID: 39938620 - Emerging Classes of Small Non-Coding RNAs With Potential Implications in Diabetes and Associated Metabolic Disorders.
Jacovetti C, Bayazit MB, Regazzi R. Jacovetti C, et al. Front Endocrinol (Lausanne). 2021 May 10;12:670719. doi: 10.3389/fendo.2021.670719. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34040585 Free PMC article. Review. - Nucleotide Modifications Decrease Innate Immune Response Induced by Synthetic Analogs of snRNAs and snoRNAs.
Stepanov G, Zhuravlev E, Shender V, Nushtaeva A, Balakhonova E, Mozhaeva E, Kasakin M, Koval V, Lomzov A, Pavlyukov M, Malyants I, Zhorov M, Kabilova T, Chernolovskaya E, Govorun V, Kuligina E, Semenov D, Richter V. Stepanov G, et al. Genes (Basel). 2018 Nov 2;9(11):531. doi: 10.3390/genes9110531. Genes (Basel). 2018. PMID: 30400232 Free PMC article.
References
- Kiss T., Fayet E., Jády B. E., Richard P., Weber M. (2006) Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 71, 407–417 - PubMed
- Matera A. G., Terns R. M., Terns M. P. (2007) Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220 - PubMed
- Kiss-László Z., Henry Y., Bachellerie J. P., Caizergues-Ferrer M., Kiss T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 85, 1077–1088 - PubMed
- Ganot P., Bortolin M. L., Kiss T. (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 89, 799–809 - PubMed
- Tollervey D., Kiss T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9, 337–342 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HL07081/HL/NHLBI NIH HHS/United States
- R01 DK064989/DK/NIDDK NIH HHS/United States
- T32 HL007081/HL/NHLBI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- K08 HL114889/HL/NHLBI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous