Salmonella Engages Host MicroRNAs To Modulate SUMOylation: a New Arsenal for Intracellular Survival - PubMed (original) (raw)
Salmonella Engages Host MicroRNAs To Modulate SUMOylation: a New Arsenal for Intracellular Survival
Smriti Verma et al. Mol Cell Biol. 2015.
Abstract
Posttranslational modifications (PTMs) can alter many fundamental properties of a protein. One or combinations of them have been known to regulate the dynamics of many cellular pathways and consequently regulate all vital processes. Understandably, pathogens have evolved sophisticated strategies to subvert these mechanisms to achieve instantaneous control over host functions. Here, we present the first report of modulation by intestinal pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) of host SUMOylation, a PTM pathway central to all fundamental cellular processes. Both in cell culture and in a mouse model, we observed that S. Typhimurium infection led to a dynamic SUMO-conjugated proteome alteration. The intracellular survival of S. Typhimurium was dependent on SUMO status as revealed by reduced infection and Salmonella-induced filaments (SIFs) in SUMO-upregulated cells. S. Typhimurium-dependent SUMO modulation was seen as a result of depletion of crucial SUMO pathway enzymes Ubc-9 and PIAS1, at both the protein and the transcript levels. Mechanistically, depletion of Ubc-9 relied on upregulation of small noncoding RNAs miR30c and miR30e during S. Typhimurium infection. This was necessary and sufficient for both down-modulation of Ubc-9 and a successful infection. Thus, we demonstrate a novel strategy of pathogen-mediated perturbation of host SUMOylation, an integral mechanism underlying S. Typhimurium infection and intracellular survival.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
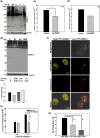
FIG 1
Salmonella infection leads to alteration in levels of SUMO-conjugated proteins in vitro and in vivo. (A and B) Comparison of cellular global SUMOylation profiling (CGSP) from uninfected human intestinal epithelial cells, HCT-8 (Ctrl), with those infected with S. Typhimurium SL1344 (ST) for 4 h. Lysates were analyzed by immunoblotting for SUMO1 (A) and SUMO2/3 (B) levels. GAPDH was used for normalization. The arrow indicates free SUMO. (C and D) CGSP assays were carried out for 4 h, and the lysates were immunoblotted using SUMO1 in J774A.2 macrophages (C) or HeLa cells (D). Numbers to the left are molecular masses in kilodaltons (also in panels F, G, I, and L). GAPDH was used for normalization. (E) Gross morphology of ceca of mice infected with S. Typhimurium (bottom) or untreated controls (Ctrl). (F and G) Immunoblot analysis of lysates prepared from mouse epithelial cells isolated from the proximal colon and probed for SUMO1 (F) and SUMO2/3 (G). Beta-actin was used for normalization. (H) Quantitative real-time PCR was carried out for all six known SENPs upon infection with S. Typhimurium for 4 h. GAPDH and 18S rRNA were used for normalization. (I) A CGSP assay was carried out in the presence of MG132, and the blot was probed for SUMO1. GAPDH was used for normalization. (J) SUMO1-conjugated and non-SUMOylated fractions of RANGAP1 over different time points of infection. GAPDH was used for normalization. (K) Samples immunoprecipitated for SUMO1- and SUMO2-conjugated proteins were probed with anti-PPARγ (top panel) and anti-RelA (bottom panel) antibodies. The asterisk indicates the SUMO-conjugated forms (J and K). Immunoprecipitation (IP) was also carried out using isotype control (IgG control). (L) Input lysates were probed with respective antibodies as well as GAPDH for normalization.
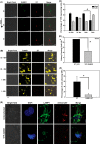
FIG 2
S. Typhimurium-mediated alteration of SUMOylation is a dynamic and active process that affects intracellular growth of the bacterium. (A) A CGSP assay was performed for the indicated time of infection, and the lysate was blotted for SUMO1. (B) CGSP assay upon 4-h infection with S. Typhimurium (ST), heat-killed S. Typhimurium (HKS), and E. coli (Ec). GAPDH was used for normalization. Means ± standard errors of the means of densitometric analysis from three independent experiments have been indicated as well as plotted. Representative blots from two to three independent experiments are depicted. (C) Gentamicin protection assays (GPAs) were performed after S. Typhimurium infection in HCT-8 cells transfected with either pEYFP vector control plasmid (VC) or pEYFP-SUMO1, and CFU scores were plotted. Efficient transfection was confirmed by YFP fluorescence imaging and immunoblot analysis (data not shown). Means ± standard errors of the means from three independent experiments are shown. Statistical analysis was carried out using Student's t test. The asterisk indicates a P value of ≤0.05. (D and E) HCT-8 cells overexpressing SUMO1 via the pCDNAHis-SUMO1 construct (D) and SUMO2 via the pCDNA-HA-SUMO2 construct (E) were infected for 4 h with S. Typhimurium, GPAs were performed, and CFU were plotted. (F) HCT-8 cells were transfected with pEYFP-SUMO1 plasmids followed by infection with mCherry-labeled S. Typhimurium for 4 h, and immunofluorescence confocal imaging was carried out. Cells with high (left panels) and low (right panels) YFP fluorescence were analyzed for bacterial load. Representative images of three independent experiments are shown. (G) Quantitative representation (means ± standard errors of the means from three independent experiments) of number of S. Typhimurium bacteria per cell plotted for low- and high-YFP-SUMO1-expressing as well as nonexpressing cells. Statistical analysis was carried out using Student's t test. The number sign indicates a P value of ≤0.01.
FIG 3
S. Typhimurium modulates the SUMOylation status of the host for SCV maturation, thus affecting long-term intracellular growth. (A) Time-lapse confocal imaging was carried out at 2 h, 3 h, and 4 h postinfection in HCT-8 cells overexpressing pEYFP-SUMO1 upon infection with mCherry-labeled S. Typhimurium (ST). (B) Confocal microscopy was carried out before and 4 h after infection with mCherry-labeled S. Typhimurium of HCT-8 cells overexpressing YFP-SUMO1. Bar, 50 μm. Representative images of three or more independent experiments are shown. (C) Quantitative representation (means ± standard errors of the means from three independent experiments, depicted in panel B) of number of high-YFP-expressing (intense and all over the cell) and low-YFP-expressing (mild and localized only to the nucleus) HCT-8 cells transfected with pEYFP-SUMO1, before and after infection with S. Typhimurium for 4 h. I, infected; UI, uninfected. Statistical analysis was carried out using Student's t test; #, P ≤ 0.01. (D) HCT-8 cells transfected with pEYFP vector control (VC) plasmid or pEYFP-SUMO1 were infected with S. Typhimurium for 24 h, and CFU were plotted. Mean values ± standard errors of the means from three independent experiments are shown. Statistical analysis was carried out using Student's t test; the asterisk indicates a P value of ≤0.05. (E) Confocal microscopic analysis of HeLa cells with (bottom panel, YFP-SUMO1) or without (top panel, untransfected) SUMO1 upregulation infected with mCherry-labeled S. Typhimurium for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs) and Salmonella-induced filaments (SIFs). Bar, 5 μm. (F) Quantitative representation (means ± standard errors of the means from three independent experiments) of percentages of cells displaying intact SIFs. Statistical analysis was carried out using Student's t test. The asterisk indicates a P value of ≤0.05.
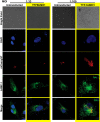
FIG 4
SUMOylation status of host affects SIF formation. Confocal microscopic analysis of HeLa cells with (YFP-SUMO1) or without (Untransfected) SUMO1 upregulation infected with different multiplicities of infection (MOIs) of mCherry-labeled S. Typhimurium for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs) and Salmonella-induced filaments (SIFs). Bar, 5 μm.
FIG 5
Salmonella targets multiple genes of SUMOylation machinery. (A to D) Immunoblot analysis of HCT-8 cells with or without infection with S. Typhimurium at different time points, probing for Sae2 (A), Ubc-9 (B), PIAS1 (C), or PIASy (D). GAPDH was used for normalization. Representative blots of three or more independent experiments are shown. Means ± standard errors of the means of densitometric analysis from three or more independent experiments are shown numerically (middle) as well as graphically (bottom). (E) A CGSP assay was carried out for 4 h, and the lysates were immunoblotted for Ubc-9, in the J774A.2 macrophage cell line (left) or HeLa cells (right). GAPDH was used for normalization. Numbers at left in panels E to G are molecular masses in kilodaltons. (F and G) Immunoblot analysis of lysates prepared from mouse epithelial cells isolated from the proximal colon and probed for Ubc-9 (F) and PIAS1 (G). Beta-actin was used for normalization. Means ± standard errors of the means of densitometric analysis from three or more independent experiments are shown numerically (middle) as well as graphically (bottom). (H) mRNA expression levels as represented by relative fold differences against uninfected control cells were plotted for the indicated genes: the Ubc-9, PIAS1, Sae2, and NF-κB genes. GAPDH and 18S rRNA were used for normalization.
FIG 6
Downregulation of Ubc-9 is sufficient to alter S. Typhimurium infection. (A and C) Immunoblot analysis of HCT-8 cells with or without downregulation of Ubc-9 (A) and PIAS1 (C) using siRNA and either infected with S. Typhimurium or left untreated. Numbers at left in panels A, C, and E are molecular masses in kilodaltons). GAPDH was used for normalization. Representative blots from three or more independent experiments are depicted. (B and D) GPA was performed in infected cells that were downregulated for either Ubc-9 (B) or PIAS1 (D). CFU were plotted after 2 h or 7 h of infection. Statistical analysis was carried out using Student's t test. The asterisk indicates a P value of ≤0.05, and the number sign indicates a P value of ≤0.01. (E) Immunoblot analysis of HCT-8 cells infected with S. Typhimurium, upon downregulation of Ubc-9 with siRNA (UKD) and upregulation of SUMO1 via pEYFP-SUMO1 (YS) or pCDNAHis-SUMO1 (HS). (F) Confocal microscopic analysis of HCT-8 cells with or without downregulation of Ubc-9 using siRNA, infected with mCherry-labeled S. Typhimurium (mST) for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs; arrow) and Salmonella-induced filaments (SIFs). Representative images of three independent experiments are depicted. Bar, 5 μm.
FIG 7
Mechanism of Ubc-9 downregulation. Mechanism of Ubc-9 downregulation was assessed via inhibitors of proteasome (MG132) (A), protein synthesis (cycloheximide) (B), and transcription (actinomycin D [AcT]) (C). Representative blots from three or more independent experiments are shown. Means ± standard errors of densitometric analysis results from three independent experiments have been indicated as well as plotted. Numbers at left of blots are molecular masses in kilodaltons.
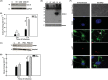
FIG 8
Salmonella engages members of the miR30 family to downregulate Ubc-9. (A) Quantitative real-time PCR analysis of mature miR30c and miR30e upon S. Typhimurium infection at different time points. SnoRNAs SNORD61 and RNU6 were used for normalization. A representative experiment of three replicates is depicted. (B) Immunoblot analysis of Ubc-9 was carried out in cells transfected with 30 nM concentrations of the mimics of miR30c and miR30e. GAPDH was used for normalization. Numbers at left of panels B and C are molecular masses in kilodaltons. (C) Ubc-9 levels were assessed in HCT-8 cells upon infection with S. Typhimurium with or without inhibitors of the miRNAs miR30c and miR30e. GAPDH was used for normalization. (D and E) GPA to assess bacterial load was performed after S. Typhimurium infection in HCT-8 cells transfected with mimics (D) or inhibitors (E) of miR30c and miR30e. CFU were plotted, and statistical analyses were carried out using Student's t test. The asterisk indicates a P value of ≤0.05, and the number sign indicates a P value of ≤0.01.
FIG 9
Inhibitors of miRNAs affect SIF formation. (A) Confocal microscopic analysis of control HeLa cells (middle panel) and HeLa cells transfected with inhibitors of miR30c (left panel) and miR30e (right panel) that were infected with mCherry-labeled S. Typhimurium for 7 h followed by immunostaining for LAMP1 to visualize the Salmonella-containing vacuoles (SCVs) and Salmonella-induced filaments (SIFs; arrows). Bars, 5 μm. (B) Quantitative representation (means ± standard errors of the means from three independent experiments) of percentages of cells displaying intact SIFs. Statistical analysis was carried out using Student's t test. The number sign indicates a P value of ≤0.01.
Similar articles
- A SUMOylation-dependent switch of RAB7 governs intracellular life and pathogenesis of Salmonella Typhimurium.
Mohapatra G, Gaur P, Mujagond P, Singh M, Rana S, Pratap S, Kaur N, Verma S, Krishnan V, Singh N, Srikanth CV. Mohapatra G, et al. J Cell Sci. 2019 Jan 11;132(1):jcs222612. doi: 10.1242/jcs.222612. J Cell Sci. 2019. PMID: 30510112 - Sumoylation as an Integral Mechanism in Bacterial Infection and Disease Progression.
Srikanth CV, Verma S. Srikanth CV, et al. Adv Exp Med Biol. 2017;963:389-408. doi: 10.1007/978-3-319-50044-7_22. Adv Exp Med Biol. 2017. PMID: 28197924 Review. - Bidirectional regulation between AP-1 and SUMOylation pathway genes modulates inflammatory signaling during Salmonella infection.
Kumar P, Soory A, Mustfa SA, Sarmah DT, Devvanshi H, Chatterjee S, Bossis G, Ratnaparkhi GS, Srikanth CV. Kumar P, et al. J Cell Sci. 2022 Aug 15;135(16):jcs260096. doi: 10.1242/jcs.260096. Epub 2022 Aug 19. J Cell Sci. 2022. PMID: 35904007 - SUMO pathway components as possible cancer biomarkers.
Mattoscio D, Chiocca S. Mattoscio D, et al. Future Oncol. 2015;11(11):1599-610. doi: 10.2217/fon.15.41. Future Oncol. 2015. PMID: 26043214 Review. - Crohn's Disease-Associated Adherent-Invasive Escherichia coli Manipulate Host Autophagy by Impairing SUMOylation.
Dalmasso G, Nguyen HTT, Faïs T, Massier S, Barnich N, Delmas J, Bonnet R. Dalmasso G, et al. Cells. 2019 Jan 9;8(1):35. doi: 10.3390/cells8010035. Cells. 2019. PMID: 30634511 Free PMC article.
Cited by
- SUMOylation of Jun fine-tunes the Drosophila gut immune response.
Soory A, Ratnaparkhi GS. Soory A, et al. PLoS Pathog. 2022 Mar 7;18(3):e1010356. doi: 10.1371/journal.ppat.1010356. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35255103 Free PMC article. - Emerging role of protein modification in inflammatory bowel disease.
Wang G, Yuan J, Luo J, Ocansey DKW, Zhang X, Qian H, Xu W, Mao F. Wang G, et al. J Zhejiang Univ Sci B. 2022 Mar 15;23(3):173-188. doi: 10.1631/jzus.B2100114. J Zhejiang Univ Sci B. 2022. PMID: 35261214 Free PMC article. Review. - Extracellular vesicles of trypomastigotes of Trypanosoma cruzi induce changes in ubiquitin-related processes, cell-signaling pathways and apoptosis.
Cornet-Gomez A, Retana Moreira L, Kronenberger T, Osuna A. Cornet-Gomez A, et al. Sci Rep. 2023 May 10;13(1):7618. doi: 10.1038/s41598-023-34820-6. Sci Rep. 2023. PMID: 37165081 Free PMC article. - MicroRNA-30e-5p Regulates SOCS1 and SOCS3 During Bacterial Infection.
Mishra R, Krishnamoorthy P, Kumar H. Mishra R, et al. Front Cell Infect Microbiol. 2021 Jan 27;10:604016. doi: 10.3389/fcimb.2020.604016. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33585275 Free PMC article. - Leishmania amazonensis sabotages host cell SUMOylation for intracellular survival.
Okuda K, Silva Costa Franco MM, Yasunaga A, Gazzinelli R, Rabinovitch M, Cherry S, Silverman N. Okuda K, et al. iScience. 2022 Aug 13;25(9):104909. doi: 10.1016/j.isci.2022.104909. eCollection 2022 Sep 16. iScience. 2022. PMID: 36060064 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous