Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes - PubMed (original) (raw)
doi: 10.1038/ng.3355. Epub 2015 Jul 13.
Daryle J DePianto 1, Justin T Jacob 1, Minerva C Han 1, Byung-Min Chung 1, Adriana S Batazzi 1, Brian G Poll 1, Yajuan Guo 1, Jingnan Han 1, SuFey Ong 2, Wenxin Zheng 1, Janis M Taube 3, Daniela Čiháková 4, Fengyi Wan 5, Pierre A Coulombe 6
Affiliations
- PMID: 26168014
- PMCID: PMC4520766
- DOI: 10.1038/ng.3355
Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes
Ryan P Hobbs et al. Nat Genet. 2015 Aug.
Abstract
Expression of the intermediate filament protein keratin 17 (K17) is robustly upregulated in inflammatory skin diseases and in many tumors originating in stratified and pseudostratified epithelia. We report that autoimmune regulator (Aire), a transcriptional regulator, is inducibly expressed in human and mouse tumor keratinocytes in a K17-dependent manner and is required for timely onset of Gli2-induced skin tumorigenesis in mice. The induction of Aire mRNA in keratinocytes depends on a functional interaction between K17 and the heterogeneous nuclear ribonucleoprotein hnRNP K. Further, K17 colocalizes with Aire protein in the nucleus of tumor-prone keratinocytes, and each factor is bound to a specific promoter region featuring an NF-κB consensus sequence in a relevant subset of K17- and Aire-dependent proinflammatory genes. These findings provide radically new insight into keratin intermediate filament and Aire function, along with a molecular basis for the K17-dependent amplification of inflammatory and immune responses in diseased epithelia.
Figures
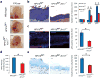
Figure 1. Loss of Krt17 attenuates _HPV16tg_-induced skin tumorigenesis
a) Macroscopic images of P70 ears from wild-type, HPV16tg/+, or HPV16tg/+;Krt17-/- mice. Scale bars = 1 cm. b) H&E staining of P70 mouse ear tissue sections and quantitation of average epidermal thickness at P20 (n = 51), P40 (n = 54), and P70 (n = 28) across genotypes. n = number of biological replicates. Scale bars = 20 μm. Error bars are s.e.m. c) Immunostaining and quantitation (n = 6 biological replicates) of phospho-Histone H3 (red) in P70 mouse ear tissue sections. Hoescht DNA stain (blue). Scale bars = 20 μm. Error bars are s.e.m. d) Myeloperoxidase (MPO) activity assay for neutrophil activation (n = 3 biological replicates, each with 4 techinical replicates). Error bars are s.e.m. e) Toluidine blue staining and quantitation (n = 6 biological replicates) of mast cells in P70 ear tissue sections. *p<0.05. Scale bars = 20 μm. Error bars are s.e.m.
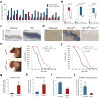
Figure 2. Aire expression, function, and regulation in skin tumor keratinocytes
a) Normalized expression for 18 of 19 gene transcripts common to A431 (red, n = 5 biological replicates) and HPV16tg/+ (blue, n = 6 biological replicates) paradigms. Error bars are s.e.m. b) Normalized expression for Aire (the 19th gene target) transcript levels in A431 (red), HPV16tg/+ (blue) and Gli2tg/+ (green) paradigms with (solid bars) or without (open bars) Krt17 expression. Error bars are s.e.m. c) Aire RNA in situ hybridization (ISH) in wild-type FVB/N mouse ear tissue sections treated with acetone or TPA. Scale bars = 10 μm. d) Aire ISH in ear tissue sections from age-matched wild-type, HPV16tg/+, and HPV16tg/+;Krt17-/- mice. Scale bars = 20 μm. e) P80 mouse ears from Gli2tg/+ and Gli2tg/+;Aire-/- littermates. Scale bars = 1 cm. Graph depicts percent Gli2tg/+;Aire-/- (n = 9) mice lesion-free over time relative to Gli2tg/+ littermates (n = 6). f) Graph depicts percent of Gli2tg/+;Krt17-/- mice (n = 22) lesion-free over time relative to Gli2tg/+ littermates (n = 15). g-h) Enrichment of Aire transcript with hnRNP K immunoprecipitation (RNA-IP) relative to IgG control in A431 keratinocytes (n = 9 biological replicates) and Gli2tg/+ keratinocytes in primary culture (n = 9 biological replicates). Error bars are s.e.m. i) TPA-induced Aire transcript levels in A431 keratinocytes expressing non-silencing (NS) or hnRNPK-targeting siRNA oligos (n = 7 biological replicates). Error bars are s.e.m. j) TPA-induced Aire transcript levels in A431 keratinocytes overexpressing hnRNP K while stably expressing shRNA targeting Krt17 (sh_Krt17_), relative to scrambled (SCR) sequence (n = 3 biological replicates). Error bars are s.e.m.
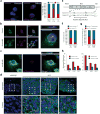
Figure 3. K17 regulates Aire subnuclear distribution and localizes to the nucleus to promote gene expression
a) Apotome-acquired images of subnuclear distribution of mCherry-Aire in A431 keratinocytes. Graph indicates percentage of cells showing punctate (blue) or diffuse (red) pattern following TPA treatment, relative to DMSO. sh_Krt17_, cells stably expressing shRNA targeting Krt17. n = number of cells analyzed. b) Single-plane confocal images of control- or leptomycin (LMB)-treated A431 keratinocytes immunostained for K17 and IκBα (positive control for LMB treatment). Inset at right highlights K17-positive nuclear punctae (arrows) and includes z-plane image. c) Same as in 3b, except A431 cells were transfected with mCherry-Aire prior to treatment and K17 immunostaining. Images in a, b, and c are representative from 15, 10, and 5 distinct experiments, respectively. Scale bars = 5 μM (a-c), 1 μm (z-planes). d) Single-plane confocal images of K17 immunostaining (or rabbit IgG control) in tissue sections of human skin basal cell carcinoma. Bottom frames, 3× digital zooms of boxed regions in top row. Arrows denote K17-positive nuclear punctae. Scale bars = 20 μm. e) Schematic of keratin protein highlighting a conserved, predicted, bipartite nuclear localization sequence (NLS) (in bold letters). Asterisk denotes Lys399. f) Graph depicting percent of HeLa cells with nuclear punctae positive for GFP-K17, wild-type (WT) or K399A (NLS mutant), as observed by confocal microscopy. n = number of cells. g) Graph depicting percent of A431 cells where mCherry-Aire punctae colocalize with GFP-K17 WT or GFP-K17 K399A. n = number of mCherry-Aire positive cells counted. h) Normalized expression of TPA-induced target gene transcript levels in A431 Krt17 null keratinocytes transfected with GFP-K17 WT or GFP-K17 K399A, either human (left) or mouse (right) species. n = three biological replicates. Error bars are s.e.m.
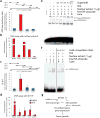
Figure 4. K17 and Aire associate with target gene promoter regions
a) Representative ChIP assay from A431 keratinocytes depicting enrichment of a specific 5′upstream sequence of CXCL11 transcript following mCherry-Aire immunoprecipitation, relative to unfused mCherry. Schematic of the 5′upstream region for Cxcl11 with locations of qRT-PCR primer sets is provided. b) Summary of ChIP assays depicting enrichment of single DNA segments within the 5′upstream sequence of select genes with mCherry-Aire IP relative to unfused mCherry. c) Summary of ChIP assays using K17 IP, relative to pre-immune sera (PIS) control, depicting TPA-induced enrichment of the same 5′upstream sequence of the CXCL11 transcript as in Fig. 4a. n = seven biological replicates. Error bars are s.e.m. d) Summary of ChIP assays using K17 IP, relative to pre-immune sera control, depicting the TPA-induced enrichment of single DNA segments within the 5′upstream sequence of select genes. n=7 biological replicates. Error bars are s.e.m. e) EMSA analysis of radiolabeled NF-κB consensus oligonucleotides using nuclear extracts from A431 keratinocytes treated with TPA, relative to DMSO control. Supershift analysis was conducted with antibodies against p65 (lane 6) and K17 (lane 7), relative to anti-IgG (lane 4) and PIS (lane 5) controls. Image represents 1 of 5 biological replicates. f) Cold competition EMSA analysis using nuclear extracts and radiolabeled oligonucleotide as in e) with inclusion of 50-fold excess non-labeled oligonucleotide (lanes 4-5). Image represents 1 of 3 biological replicates. g) K17 regulates Aire at both the transcript and protein levels to promote inflammatory gene expression and skin tumorigenesis.
Similar articles
- Regulation of C-X-C chemokine gene expression by keratin 17 and hnRNP K in skin tumor keratinocytes.
Chung BM, Arutyunov A, Ilagan E, Yao N, Wills-Karp M, Coulombe PA. Chung BM, et al. J Cell Biol. 2015 Mar 2;208(5):613-27. doi: 10.1083/jcb.201408026. Epub 2015 Feb 23. J Cell Biol. 2015. PMID: 25713416 Free PMC article. - The autoimmune regulator (AIRE), which is defective in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients, is expressed in human epidermal and follicular keratinocytes and associates with the intermediate filament protein cytokeratin 17.
Kumar V, Pedroza LA, Mace EM, Seeholzer S, Cotsarelis G, Condino-Neto A, Payne AS, Orange JS. Kumar V, et al. Am J Pathol. 2011 Mar;178(3):983-8. doi: 10.1016/j.ajpath.2010.12.007. Am J Pathol. 2011. PMID: 21356351 Free PMC article. - A role for keratin 17 during DNA damage response and tumor initiation.
Nair RR, Hsu J, Jacob JT, Pineda CM, Hobbs RP, Coulombe PA. Nair RR, et al. Proc Natl Acad Sci U S A. 2021 Mar 30;118(13):e2020150118. doi: 10.1073/pnas.2020150118. Proc Natl Acad Sci U S A. 2021. PMID: 33762306 Free PMC article. - Keratin 17 as a therapeutic target for the treatment of psoriasis.
Fu M, Wang G. Fu M, et al. J Dermatol Sci. 2012 Sep;67(3):161-5. doi: 10.1016/j.jdermsci.2012.06.008. Epub 2012 Jun 23. J Dermatol Sci. 2012. PMID: 22795618 Review. - Keratin 17 in disease pathogenesis: from cancer to dermatoses.
Yang L, Zhang S, Wang G. Yang L, et al. J Pathol. 2019 Feb;247(2):158-165. doi: 10.1002/path.5178. Epub 2018 Dec 7. J Pathol. 2019. PMID: 30306595 Review.
Cited by
- Keratins Are Going Nuclear.
Hobbs RP, Jacob JT, Coulombe PA. Hobbs RP, et al. Dev Cell. 2016 Aug 8;38(3):227-33. doi: 10.1016/j.devcel.2016.07.022. Dev Cell. 2016. PMID: 27505414 Free PMC article. - Significance of stress keratin expression in normal and diseased epithelia.
Cohen E, Johnson CN, Wasikowski R, Billi AC, Tsoi LC, Kahlenberg JM, Gudjonsson JE, Coulombe PA. Cohen E, et al. iScience. 2024 Jan 5;27(2):108805. doi: 10.1016/j.isci.2024.108805. eCollection 2024 Feb 16. iScience. 2024. PMID: 38299111 Free PMC article. - Keratin 17 covalently binds to alpha-enolase and exacerbates proliferation of keratinocytes in psoriasis.
Luo Y, Pang B, Hao J, Li Q, Qiao P, Zhang C, Bai Y, Xiao C, Chen J, Zhi D, Liu Y, Dang E, Wang G, Li B. Luo Y, et al. Int J Biol Sci. 2023 Jul 3;19(11):3395-3411. doi: 10.7150/ijbs.83141. eCollection 2023. Int J Biol Sci. 2023. PMID: 37497003 Free PMC article. - Cytoplasmic keratins couple with and maintain nuclear envelope integrity in colonic epithelial cells.
Stenvall CA, Nyström JH, Butler-Hallissey C, Jansson T, Heikkilä TRH, Adam SA, Foisner R, Goldman RD, Ridge KM, Toivola DM. Stenvall CA, et al. Mol Biol Cell. 2022 Nov 1;33(13):ar121. doi: 10.1091/mbc.E20-06-0387. Epub 2022 Aug 24. Mol Biol Cell. 2022. PMID: 36001365 Free PMC article. - Keratin 6a reorganization for ubiquitin-proteasomal processing is a direct antimicrobial response.
Chan JKL, Yuen D, Too PH, Sun Y, Willard B, Man D, Tam C. Chan JKL, et al. J Cell Biol. 2018 Feb 5;217(2):731-744. doi: 10.1083/jcb.201704186. Epub 2017 Nov 30. J Cell Biol. 2018. PMID: 29191848 Free PMC article.
References
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. - PubMed
- Jin L, Wang G. Keratin 17: a critical player in the pathogenesis of psoriasis. Med Res Rev. 2014;34:438–454. - PubMed
Methods Only References
- Reichelt J, Haase I. Establishment of spontaneously immortalized keratinocyte lines from wild-type and mutant mice. Methods Mol Biol. 2010;585:59–69. - PubMed
- Adamson KA, Pearce SH, Lamb JR, Seckl JR, Howie SE. A comparative study of mRNA and protein expression of the autoimmune regulator gene (Aire) in embryonic and adult murine tissues. J Pathol. 2004;202:180–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM111682/GM/NIGMS NIH HHS/United States
- R01 CA160255/CA/NCI NIH HHS/United States
- T32 CA009110/CA/NCI NIH HHS/United States
- GM111682/GM/NIGMS NIH HHS/United States
- AR044232/AR/NIAMS NIH HHS/United States
- CA160255/CA/NCI NIH HHS/United States
- R01 AR044232/AR/NIAMS NIH HHS/United States
- CA009110/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous