The NF45/NF90 Heterodimer Contributes to the Biogenesis of 60S Ribosomal Subunits and Influences Nucleolar Morphology - PubMed (original) (raw)
The NF45/NF90 Heterodimer Contributes to the Biogenesis of 60S Ribosomal Subunits and Influences Nucleolar Morphology
Franziska Wandrey et al. Mol Cell Biol. 2015 Oct.
Abstract
The interleukin enhancer binding factors ILF2 (NF45) and ILF3 (NF90/NF110) have been implicated in various cellular pathways, such as transcription, microRNA (miRNA) processing, DNA repair, and translation, in mammalian cells. Using tandem affinity purification, we identified human NF45 and NF90 as components of precursors to 60S (pre-60S) ribosomal subunits. NF45 and NF90 are enriched in nucleoli and cosediment with pre-60S ribosomal particles in density gradient analysis. We show that association of the NF45/NF90 heterodimer with pre-60S ribosomal particles requires the double-stranded RNA binding domains of NF90, while depletion of NF45 and NF90 by RNA interference leads to a defect in 60S biogenesis. Nucleoli of cells depleted of NF45 and NF90 have altered morphology and display a characteristic spherical shape. These effects are not due to impaired rRNA transcription or processing of the precursors to 28S rRNA. Consistent with a role of the NF45/NF90 heterodimer in nucleolar steps of 60S subunit biogenesis, downregulation of NF45 and NF90 leads to a p53 response, accompanied by induction of the cyclin-dependent kinase inhibitor p21/CIP1, which can be counteracted by depletion of RPL11. Together, these data indicate that NF45 and NF90 are novel higher-eukaryote-specific factors required for the maturation of 60S ribosomal subunits.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
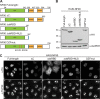
FIG 1
NF45 and NF90, but not NF110, are components of pre-60S ribosomal particles. (A) HEK293 cells expressing ZNF622-StHA were used for TAP. The purified proteins were analyzed by SDS-PAGE, followed by Coomassie staining and mass spectrometry analysis of the excised bands (see Table S1 in the supplemental material). The proteins detected with the highest peptide numbers are listed on the right. Gray protein names indicate proteins for which there are either no yeast homologs or yeast homologs that have not been implicated in ribosome biogenesis. TAP of ZNF622-StHA purifies pre-60S ribosomal particles as well as NF45 and NF90. (B) HeLa cell extract was centrifuged on a 10% to 45% sucrose gradient. Cell extract (Input) and gradient fractions were analyzed by Western blotting using the indicated antibodies. Note that the anti-ILF3 antibody recognizes both the NF90 and the NF110 isoforms of ILF3. Fractions containing 40S and 60S particles are indicated at the bottom. NF45 and NF90 comigrate in fractions containing 60S particles, whereas NF110 is present at the top of the gradient. (C) TAP of HEK293 cells expressing the 60S _trans_-acting factor ZNF622-StHA or the 40S _trans_-acting factor HASt-LTV1 or HASt-PNO1 was performed. Cleared cell extracts (Input) and eluted proteins (Eluate) were analyzed by Western blotting using the indicated antibodies. NF45 and NF90, but not NF110, were copurified by ZNF622 TAP but were not present in the eluate samples obtained by LTV1 or PNO1 TAP.
FIG 2
NF45 and NF90 localize to the nucleus and are enriched in nucleoli. (A) Extract from HeLa cells was fractionated, and equal volumes of total cells, cytoplasmic extract, and the pellet containing the nucleus were analyzed by Western blotting using the indicated antibodies. NF45 and NF90 were exclusively present in the nuclear fraction at steady state. (B) Localization of NF45 and NF90/NF110 in HeLa cells was analyzed by immunofluorescence with the indicated antibodies. NF45 was enriched in nucleoli, whereas NF110 was predominantly localized to the nucleoplasm. Scale bar, 20 μm. (C) HeLa cells were transiently transfected with N- and C-terminally HASt-tagged NF90 or NF110. The subcellular localization of the tagged proteins was detected by immunofluorescence using an anti-HA antibody. Nucleoli were visualized by coimmunofluorescence against RLP24. Scale bar, 20 μm. (D) Western blot analysis of cells from panel C using the indicated antibodies to monitor expression levels of transfected constructs.
FIG 3
NF90 TAP copurifies pre-60S ribosomal particles. TAP was performed using HEK293 cells expressing either HASt-NF90 or HASt-GFP (negative control) as bait. (A) Eluted proteins were analyzed by SDS-PAGE, followed by silver staining (shown) or Coomassie blue staining. Bands visible by Coomassie blue staining were excised and analyzed by mass spectrometry (see Table S2 in the supplemental material). The proteins with the highest numbers of peptides detected are indicated on the right. Gray protein names indicate proteins for which there are either no yeast homologs or yeast homologs that have not been implicated in ribosome biogenesis. Baits are marked with asterisks. (B) Western blot analysis of the TAP experiment used for panel A with the indicated antibodies. NF90 copurifies NF45 and ribosomal proteins of the 60S subunit, as well as 60S but not 40S _trans_-acting factors.
FIG 4
The dsRBDs of NF90 are required for nucleolar localization. (A) Scheme of generated NF90 truncations/mutants. Full-length NF90 possesses an N-terminal DZF domain with which it dimerizes with NF45, an NLS (blue), and two dsRBD domains. The amino acid numbers indicate the positions of domains and the length of each NF90 truncation. The two mutated amino acid residues for the DZF mutant are shown in red. (B) The N-terminally HASt-tagged NF90 constructs from panel A were transiently transfected into HeLa cells for 24 h. Cells were harvested and analyzed by Western blotting using the indicated antibodies. (C) Cells transfected as in panel B were fixed and analyzed by immunofluorescence using the indicated antibodies. Scale bar, 20 μm.
FIG 5
The dsRBDs of NF90 are required for association with pre-60S ribosomal particles. (A) TAP using HEK293 cell lines inducibly expressing the indicated HASt-tagged NF90 constructs and HASt-GFP as a negative control. Baits are marked with asterisks. (A and B) The eluates were analyzed by SDS-PAGE, followed by silver staining (A) or Western blotting (B) using the indicated antibodies. NF90 truncations lacking the dsRBDs did not copurify pre-60S ribosomal particles.
FIG 6
NF45/NF90 depletion leads to a ribosome biogenesis defect. (A) HeLa cells expressing RPL29-GFP or RPL26-GFP under a tetracycline-dependent promoter were treated with either control siRNA (si-control) or siRNAs against NF45 or NF90/NF110 for 72 h. Cells were fixed and analyzed by fluorescence microscopy. Scale bar, 20 μm. (B) Western blot analysis to control for downregulation of NF45 and NF90 in RPL29-GFP-expressing cells shown in panel A using the indicated antibodies. (C) HeLa K cells were treated with either control siRNA (si-control) or siRNAs against NF45 for 72 h. Cell extracts were separated by centrifugation on a linear 10% to 45% sucrose gradient. Cell extract (Input) and gradient fractions were analyzed by Western blotting using the indicated antibodies. Fractions containing 40S and 60S particles are indicated at the bottom. Note that binding of LSG1 to pre-60S ribosomal particles is diminished upon NF45 depletion. (D) To confirm NF45 downregulation, cell extracts shown in panel C were analyzed by Western blotting using the indicated antibodies.
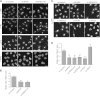
FIG 7
NF45/NF90 depletion leads to altered nucleolar morphology. (A) HeLa K cells were treated with either control siRNA (si-control) or siRNA against NF45 or NF90/NF110 for 72 h. The cells were fixed and analyzed by IF using the indicated antibodies. Wider spaces between rows separate independent experiments. Scale bar, 20 μm. (B) Quantification of nucleolar shapes of cells from three independent experiments using the anti-eIF6 readout. The error bars indicate standard deviations. Statistically significant differences from control cells, determined by a t test, are indicated (**, P ≤ 0.01). (C) HeLa cells were transfected with the indicated siRNAs for 72 h and analyzed by immunofluorescence using an antibody against eIF6. Scale bar, 20 μm. (D) Quantification of nucleolar shapes from three independent experiments analogous to those used for panel B (**, P ≤ 0.01; *, P ≤ 0.05).
FIG 8
Depletion of NF45 and NF90 does not cause cell cycle arrest in HeLa cells but leads to RPL11-dependent p21 induction. (A) HeLa cells were treated with the indicated siRNAs for 72 h and analyzed by Western blotting with the indicated antibodies. (B) HeLa cells were treated with the indicated siRNAs for 72 h and analyzed by flow cytometry. The results of three independent experiments were quantified, and the percentage of cells in G1 for each condition is shown with the standard deviation. A t test was performed to determine significant differences (ns, not significant). (C) Flow cytometry histograms of one representative experiment shown in panel B. Cell cycle stages are indicated in the first histogram. (D) Immunofluorescence analysis of one of the experiments used in panel B using an antibody against eIF6. Scale bar, 20 μm. (E) Quantification of nucleolar shapes of cells shown in panel D. For each condition, >300 nucleoli were analyzed. The error bars show standard deviations, and a t test was performed to determine significant differences (***, P < 0.001). (F) U2OS cells were treated with the indicated siRNAs for 72 h and analyzed by Western blotting with the indicated antibodies.
Similar articles
- Inducible expression of immediate early genes is regulated through dynamic chromatin association by NF45/ILF2 and NF90/NF110/ILF3.
Wu TH, Shi L, Lowe AW, Nicolls MR, Kao PN. Wu TH, et al. PLoS One. 2019 Apr 25;14(4):e0216042. doi: 10.1371/journal.pone.0216042. eCollection 2019. PLoS One. 2019. PMID: 31022259 Free PMC article. - Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control.
Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, Li H, Lee CG, Pe'ery T, Mathews MB. Guan D, et al. Mol Cell Biol. 2008 Jul;28(14):4629-41. doi: 10.1128/MCB.00120-08. Epub 2008 May 5. Mol Cell Biol. 2008. PMID: 18458058 Free PMC article. - Induction of p53, p21 and apoptosis by silencing the NF90/NF45 complex in human papilloma virus-transformed cervical carcinoma cells.
Shamanna RA, Hoque M, Pe'ery T, Mathews MB. Shamanna RA, et al. Oncogene. 2013 Oct 24;32(43):5176-85. doi: 10.1038/onc.2012.533. Epub 2012 Dec 3. Oncogene. 2013. PMID: 23208500 Free PMC article. - SUMO routes ribosome maturation.
Finkbeiner E, Haindl M, Raman N, Muller S. Finkbeiner E, et al. Nucleus. 2011 Nov-Dec;2(6):527-32. doi: 10.4161/nucl.2.6.17604. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064470 Review. - Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.
Konikkat S, Woolford JL Jr. Konikkat S, et al. Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review.
Cited by
- A novel role for the peptidyl-prolyl cis-trans isomerase Cyclophilin A in DNA-repair following replication fork stalling via the MRE11-RAD50-NBS1 complex.
Bedir M, Outwin E, Colnaghi R, Bassett L, Abramowicz I, O'Driscoll M. Bedir M, et al. EMBO Rep. 2024 Aug;25(8):3432-3455. doi: 10.1038/s44319-024-00184-9. Epub 2024 Jun 28. EMBO Rep. 2024. PMID: 38943005 Free PMC article. - Inducible expression of immediate early genes is regulated through dynamic chromatin association by NF45/ILF2 and NF90/NF110/ILF3.
Wu TH, Shi L, Lowe AW, Nicolls MR, Kao PN. Wu TH, et al. PLoS One. 2019 Apr 25;14(4):e0216042. doi: 10.1371/journal.pone.0216042. eCollection 2019. PLoS One. 2019. PMID: 31022259 Free PMC article. - Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins.
Van Nostrand EL, Pratt GA, Yee BA, Wheeler EC, Blue SM, Mueller J, Park SS, Garcia KE, Gelboin-Burkhart C, Nguyen TB, Rabano I, Stanton R, Sundararaman B, Wang R, Fu XD, Graveley BR, Yeo GW. Van Nostrand EL, et al. Genome Biol. 2020 Apr 6;21(1):90. doi: 10.1186/s13059-020-01982-9. Genome Biol. 2020. PMID: 32252787 Free PMC article. - Tracing Eukaryotic Ribosome Biogenesis Factors Into the Archaeal Domain Sheds Light on the Evolution of Functional Complexity.
Birikmen M, Bohnsack KE, Tran V, Somayaji S, Bohnsack MT, Ebersberger I. Birikmen M, et al. Front Microbiol. 2021 Sep 16;12:739000. doi: 10.3389/fmicb.2021.739000. eCollection 2021. Front Microbiol. 2021. PMID: 34603269 Free PMC article. - Ribosome biogenesis factors-from names to functions.
Dörner K, Ruggeri C, Zemp I, Kutay U. Dörner K, et al. EMBO J. 2023 Apr 3;42(7):e112699. doi: 10.15252/embj.2022112699. Epub 2023 Feb 10. EMBO J. 2023. PMID: 36762427 Free PMC article. Review.
References
- Corthésy B, Kao PN. 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem 269:20682–20690. - PubMed
- Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthésy B. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem 269:20691–20699. - PubMed
- Marcoulatos P, Koussidis G, Mamuris Z, Velissariou V, Vamvakopoulos NC. 1996. Mapping interleukin enhancer binding factor 2 gene (ILF2) to human chromosome 1 (1q11-qter and 1p11-p12) by polymerase chain reaction amplification of human-rodent somatic cell hybrid DNA templates. J Interferon Cytokine Res 16:1035–1038. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous