Topography of the Human Papillomavirus Minor Capsid Protein L2 during Vesicular Trafficking of Infectious Entry - PubMed (original) (raw)
Topography of the Human Papillomavirus Minor Capsid Protein L2 during Vesicular Trafficking of Infectious Entry
Stephen DiGiuseppe et al. J Virol. 2015 Oct.
Abstract
The human papillomavirus (HPV) capsid is composed of the major capsid protein L1 and the minor capsid protein L2. During entry, the HPV capsid undergoes numerous conformational changes that result in endosomal uptake and subsequent trafficking of the L2 protein in complex with the viral DNA to the trans-Golgi network. To facilitate this transport, the L2 protein harbors a number of putative motifs that, if capable of direct interaction, would interact with cytosolic host cell factors. These data imply that a portion of L2 becomes cytosolic during infection. Using a low concentration of digitonin to selectively permeabilize the plasma membrane of infected cells, we mapped the topography of the L2 protein during infection. We observed that epitopes within amino acid residues 64 to 81 and 163 to 170 and a C-terminal tag of HPV16 L2 are exposed on the cytosolic side of intracellular membranes, whereas an epitope within residues 20 to 38, which are upstream of a putative transmembrane region, is luminal. Corroborating these findings, we also found that L2 protein is sensitive to trypsin digestion during infection. These data demonstrate that the majority of the L2 protein becomes accessible on the cytosolic side of intracellular membranes in order to interact with cytosolic factors to facilitate vesicular trafficking.
Importance: In order to complete infectious entry, nonenveloped viruses have to pass cellular membranes. This is often achieved through the viral capsid protein associating with or integrating into intracellular membrane. Here, we determine the topography of HPV L2 protein in the endocytic vesicular compartment, suggesting that L2 becomes a transmembrane protein with a short luminal portion and with the majority facing the cytosolic side for interaction with host cell transport factors.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
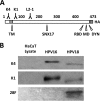
Characterization of L2-specific monoclonal antibodies. (A) Schematic of the L2 protein with labeled key features. Numbers refer to amino acid positions of HPV16 L2: K4, MAb K4 epitope (aa 20 to 38); TM, N-terminal putative transmembrane region (aa 45 to 67); K1, MAb K1 epitope (aa 64 to 81); L2-1, MAb 33L2-1 epitope (aa 163 to 170); SNX17, sorting nexin 17 binding domain (aa 245 to 257); RBD, retromer binding domain (aa 446 to 455); MD, membrane-destabilizing domain (aa 454 to 473); DYN, dynein binding domain (aa 456 to 461); HA, C-terminal HA epitope. (B) Western blot using L2-specific antibodies (listed on the left side) to detect L2 protein from purified HPV16 and HPV18 pseudoviruses. To exclude cross-reactivity, HaCaT whole-cell extracts were run as controls.
FIG 2
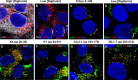
Detection of L2 protein during infection. HaCaT cells were infected with HPV16 pseudovirus for 24 h and analyzed by immunofluorescence microscopy. The L2 protein (detected by K4, K1, or 33L2-1), EdU-labeled pseudogenome, TGN marker TGN46, and nucleus (DAPI) were stained following a Click-iT reaction. Intracellular L2 was detected using MAb K4 (aa 20 to 38) (A), MAb K1 (aa 64 to 81) (D), and MAb 33L2-1 (aa 163 to 170) (G). (B, E, and H) Colocalization was confirmed by analyzing the line profile of individual EdU puncta. (C, F, and I) Mock-infected HaCaT cells served as controls to assess the amount of background signal from each of the MAbs used. Arrowheads indicate the tri-color (white) colocalization of the L2-specific antibodies with TGN46 and EdU signals.
FIG 3
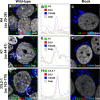
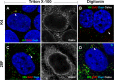
Low concentration of digitonin selectively permeabilizes the plasma membrane. (A and B) HaCaT cells were grown for 24 h and then fixed and permeabilized with either 1 mg/ml (A) or 5 μg/ml (B) digitonin. Lamp1, cytosol-facing late endosome marker Lamp1; calnexin (Calnx), luminal endoplasmic reticulum marker; DAPI, nuclear marker. (C and D) HaCaT cells were grown for 24 h and then fixed and permeabilized with either 0.5% Triton X-100 or 5 μg/ml digitonin. p230, cytosol-facing TGN marker; TGN46, luminal TGN marker; DAPI, nuclear marker. (E to H) HaCaT cells were infected with HPV16 pseudovirus for 18 h in the presence of 1 μM bafilomycin A1. Cells were fixed, permeabilized using 5 μg/ml digitonin, and stained using L2-specific antibodies (K4, K1, and 33L2-1) or an L1-specific antibody (33L1-7) following the Click-iT reaction. Colors are coded as indicated by the labels on the figure.
FIG 4
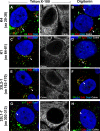
Regions of 16L2 downstream of the N-terminal putative transmembrane domain become accessible in selectively permeabilized cells. HaCaT cells were infected for 24 h with HPV16 pseudovirus. Cells were fixed and permeabilized using 0.5% Triton X-100 (A, C, E, and G) or 5 μg/ml digitonin (B, D, F, and H) and stained using L2-specific MAbs (K4, K1, and 33L2-1) or an L1-specific MAb (33L1-7). The ER was stained using MAb against the luminal ER marker calnexin (Calnx). Cells were fixed again and permeabilized using 0.5% Triton X-100, followed by treatment with the Click-iT reaction mixture to detect EdU. Note that only the L2-specific MAb K4, but not K1 and 33L2-1 MAbs, fails to detect the L2 protein adjacent to the nucleus, probably residing in the TGN (arrows), under selective permeabilization. (G and H) As a control, we also did not detect 33L1-7 staining following the Click-iT reaction adjacent to the nucleus under selective permeabilization. Colors are coded as indicated by the labels on the figure.
FIG 5
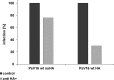
Microinjection of anti-HA antibody neutralizes infection in vivo. 293TT cells were infected with 16L2-expressing pseudovirus (PsV16 wt) or C-terminally tagged 16L2-HA-3′-expressing pseudovirus (HA) for 72 h. Cells were microinjected with purified MAb anti-HA antibody in vivo at 2 hpi using a Leica mechanical manipulator. Infectivity was quantified by counting the number of GFP-expressing cells (n > 100 cells each).
FIG 6
Regions of 18L2 downstream of the N-terminal putative transmembrane domain become accessible in selectively permeabilized cells. HaCaT cells were infected for 24 h with HPV18 pseudovirus. Cells were fixed and permeabilized using 0.5% Triton X-100 (A and C) or 5 μg/ml digitonin (B and D) and stained using L2-specific MAbs (K4 and 28F). The ER was stained using MAb against the luminal ER marker calnexin (Calnx). Cells were fixed again and permeabilized using 0.5% Triton X-100, followed by treatment with the Click-iT reaction mixture to detect EdU. Colors are coded as indicated by the labels on the figure. Note that only the L2-specific MAb K4 and not 28F fails to detect the L2 protein adjacent to the nucleus, probably residing in the TGN (arrows), under selective permeabilization.
FIG 7
The majority of the L2 protein becomes exposed within the cytoplasm during infectious entry. (A) Cytoplasm-localized puncta were selected from the experiments shown in both Fig. 4 (PSV16, HPV16 pseudovirus) and 6 (PSV18, HPV18 pseudovirus) and quantified from infected HaCaT cells treated with either 0.5% Triton X-100 (TX) or 5 μg/ml digitonin (DIG). The ratio of L2 to EdU was determined by measuring the signal strength of each channel. (B) Histograms of signal strengths for all channels from select representative puncta from the experiments shown in both Fig. 4 and 6. Note the lack of L2-specific MAb K4 signal under selective permeabilization. (C) HeLa cells were infected with 18L2-R295/8A-harboring pseudovirions for 18 h. Cells were harvested, and lysates were prepared by mechanically disrupting the plasma membranes. Whole-cell extracts were treated with or without 1 μM bafilomycin A1 and/or 0.2% trypsin and/or Triton X-100 for 1 h. Western blotting was performed, and the L2 protein was detected using a cocktail of 18L2-specific MAbs. Detection of BiP protein served as an internal control. The protein levels were quantified by measuring the pixel intensity of each band using densitometry relative to their respective control lanes. Note that the L2 protein is sensitive to trypsin, regardless of the addition of 0.5% Triton X-100. ns, not significant.
Similar articles
- A Novel PDZ Domain Interaction Mediates the Binding between Human Papillomavirus 16 L2 and Sorting Nexin 27 and Modulates Virion Trafficking.
Pim D, Broniarczyk J, Bergant M, Playford MP, Banks L. Pim D, et al. J Virol. 2015 Oct;89(20):10145-55. doi: 10.1128/JVI.01499-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202251 Free PMC article. - Phosphorylation of Human Papillomavirus Type 16 L2 Contributes to Efficient Virus Infectious Entry.
Broniarczyk J, Massimi P, Pim D, Bergant Marušič M, Myers MP, Garcea RL, Banks L. Broniarczyk J, et al. J Virol. 2019 Jun 14;93(13):e00128-19. doi: 10.1128/JVI.00128-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996086 Free PMC article. - Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection.
Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. Popa A, et al. PLoS Pathog. 2015 Feb 18;11(2):e1004699. doi: 10.1371/journal.ppat.1004699. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25693203 Free PMC article. - L2, the minor capsid protein of papillomavirus.
Wang JW, Roden RB. Wang JW, et al. Virology. 2013 Oct;445(1-2):175-86. doi: 10.1016/j.virol.2013.04.017. Epub 2013 May 17. Virology. 2013. PMID: 23689062 Free PMC article. Review. - Subcellular Trafficking of the Papillomavirus Genome during Initial Infection: The Remarkable Abilities of Minor Capsid Protein L2.
Campos SK. Campos SK. Viruses. 2017 Dec 3;9(12):370. doi: 10.3390/v9120370. Viruses. 2017. PMID: 29207511 Free PMC article. Review.
Cited by
- A Ran-binding protein facilitates nuclear import of human papillomavirus type 16.
Lai KY, Rizzato M, Aydin I, Villalonga-Planells R, Drexler HCA, Schelhaas M. Lai KY, et al. PLoS Pathog. 2021 May 11;17(5):e1009580. doi: 10.1371/journal.ppat.1009580. eCollection 2021 May. PLoS Pathog. 2021. PMID: 33974675 Free PMC article. - Human Papillomavirus L2 Capsid Protein Stabilizes γ-Secretase during Viral Infection.
Crite M, DiMaio D. Crite M, et al. Viruses. 2022 Apr 13;14(4):804. doi: 10.3390/v14040804. Viruses. 2022. PMID: 35458534 Free PMC article. - Sequence-independent activity of a predicted long disordered segment of the human papillomavirus type 16 L2 capsid protein during virus entry.
Oh C, Buckley PM, Choi J, Hierro A, DiMaio D. Oh C, et al. Proc Natl Acad Sci U S A. 2023 Oct 17;120(42):e2307721120. doi: 10.1073/pnas.2307721120. Epub 2023 Oct 11. Proc Natl Acad Sci U S A. 2023. PMID: 37819982 Free PMC article. - PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery.
Guion L, Bienkowska-Haba M, DiGiuseppe S, Florin L, Sapp M. Guion L, et al. PLoS Pathog. 2019 Feb 25;15(2):e1007590. doi: 10.1371/journal.ppat.1007590. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30802273 Free PMC article. - Non-enveloped virus membrane penetration: New advances leading to new insights.
Pletan ML, Tsai B. Pletan ML, et al. PLoS Pathog. 2022 Dec 8;18(12):e1010948. doi: 10.1371/journal.ppat.1010948. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36480535 Free PMC article.
References
- Finch JT, Klug A. 1965. The structure of viruses of the papilloma-polyoma type 3. Structure of rabbit papilloma virus, with an appendix on the topography of contrast in negative-staining for electron-microscopy. J Mol Biol 13:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM103433/GM/NIGMS NIH HHS/United States
- R01 AI081809/AI/NIAID NIH HHS/United States
- P20GM103433/GM/NIGMS NIH HHS/United States
- R01AI081809/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous