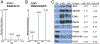Redefining the Breast Cancer Exosome Proteome by Tandem Mass Tag Quantitative Proteomics and Multivariate Cluster Analysis - PubMed (original) (raw)
Redefining the Breast Cancer Exosome Proteome by Tandem Mass Tag Quantitative Proteomics and Multivariate Cluster Analysis
David J Clark et al. Anal Chem. 2015.
Abstract
Exosomes are microvesicles of endocytic origin constitutively released by multiple cell types into the extracellular environment. With evidence that exosomes can be detected in the blood of patients with various malignancies, the development of a platform that uses exosomes as a diagnostic tool has been proposed. However, it has been difficult to truly define the exosome proteome due to the challenge of discerning contaminant proteins that may be identified via mass spectrometry using various exosome enrichment strategies. To better define the exosome proteome in breast cancer, we incorporated a combination of Tandem-Mass-Tag (TMT) quantitative proteomics approach and Support Vector Machine (SVM) cluster analysis of three conditioned media derived fractions corresponding to a 10 000g cellular debris pellet, a 100 000g crude exosome pellet, and an Optiprep enriched exosome pellet. The quantitative analysis identified 2 179 proteins in all three fractions, with known exosomal cargo proteins displaying at least a 2-fold enrichment in the exosome fraction based on the TMT protein ratios. Employing SVM cluster analysis allowed for the classification 251 proteins as "true" exosomal cargo proteins. This study provides a robust and vigorous framework for the future development of using exosomes as a potential multiprotein marker phenotyping tool that could be useful in breast cancer diagnosis and monitoring disease progression.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1.
Overview of exosome isolation, characterization, and TMT-labeling strategy to elucidate exosomal protein enrichment. (A) Differential ultracentrifugation was performed, and pellets obtained after 10 000g spin (10K pellet), 100 000g spin (100 K pellet), and post-Optiprep density gradient (Opti pellet) were retained for MS analysis. (B) Immunoblot for known exosome markers ALIX and CD63 in the recovered 1 mL Optiprep fractions. Fractions 6 and 7 were positive for the exosome markers with a corresponding density of 1.14–1.19 g/mL. (C) Electron microscopy image of negatively stained SKBR3B exosome vesicles. Bar = 100 nm. (D) Peptide digest from each fraction was TMT-labeled and mixed equally prior to LC–MS/MS analysis. Hybridization method of PQD-CID allowed for quantification of reporter ions and identification of the peptide.
Figure 2.
Peptides and proteins display differential abundance in various analysis fractions. Identified peptides from ATP synthase subunit δ (A) and CD63 (B) display differences in TMT reporter ion intensities between the three fractions. TMT reporter ions are as follows: 129–10K fraction, 130–100 K fraction, and 131-Opti fraction. (C) Western Blot analysis of individual fraction lysates reveals differential protein abundance (left). Known exosome proteins ALIX, TSG101, and CD63 as well as GSK3_β_ increased in abundance with exosome enrichment. Levels of nuclear proteins (PARP1 and Histone H3 (HISTH3A)), cytoskeletal protein _β_-actin, and the mitochondrial protein, Cytochrome C (CYC1), decreased during exosome enrichment. Reported QuantiMORE TMT ratios for each respective protein representing 100 000g ultracentrifugation (100K/10K) and Optiprep (Opti/10K) exosome enrichment (right).
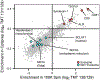
Figure 3.
Known exosome markers localize to the quadrant corresponding to 100 K and Optiprep enrichment. TMT log2 protein ratios were plotted by 100K enrichment (x-axis) against Optiprep enrichment (y-axis). Selected Exosome (red) and Nonexosome (blue) protein markers are annotated.
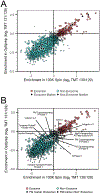
Figure 4.
SVM cluster analysis classified 241 proteins as exosomal: (A) plotted SVM cluster of all 2 179 quantified proteins from the TMT analysis. Exosome proteins (red circles) localized in the upper right quadrant, with exosome markers and nonexosome markers indicated by black circles. Proteins classified as nonexosomal are indicated by the blue squares. (B) Plotted SVM cluster with annotated plasma membrane markers indicated by black circles.
Similar articles
- [Tandem mass tag-based quantitative proteomics analysis of plasma and plasma exosomes in Parkinson's disease].
Zhao Y, Liu X, Zhang Y, Zhang J, Liu X, Yang G. Zhao Y, et al. Se Pu. 2023 Dec;41(12):1073-1083. doi: 10.3724/SP.J.1123.2022.12022. Se Pu. 2023. PMID: 38093537 Free PMC article. Chinese. - The core exosome proteome of Trichomonas vaginalis.
Ong SC, Luo HW, Cheng WH, Ku FM, Tsai CY, Huang PJ, Lee CC, Yeh YM, Lin R, Chiu CH, Tang P. Ong SC, et al. J Microbiol Immunol Infect. 2024 Apr;57(2):246-256. doi: 10.1016/j.jmii.2024.02.003. Epub 2024 Feb 16. J Microbiol Immunol Infect. 2024. PMID: 38383245 - Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients.
Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, Gascoyne P, Mulu FC, Stephens BM, Huang J, Wang H, Momin AA, Jacamo RO, Katz M, Wolff R, Javle M, Varadhachary G, Wistuba II, Hanash S, Maitra A, Alvarez H. Castillo J, et al. Ann Oncol. 2018 Jan 1;29(1):223-229. doi: 10.1093/annonc/mdx542. Ann Oncol. 2018. PMID: 29045505 Free PMC article. - Proteomics characterization of exosome cargo.
Schey KL, Luther JM, Rose KL. Schey KL, et al. Methods. 2015 Oct 1;87:75-82. doi: 10.1016/j.ymeth.2015.03.018. Epub 2015 Mar 31. Methods. 2015. PMID: 25837312 Free PMC article. Review. - The human urinary exosome as a potential metabolic effector cargo.
Bruschi M, Ravera S, Santucci L, Candiano G, Bartolucci M, Calzia D, Lavarello C, Inglese E, Petretto A, Ghiggeri G, Panfoli I. Bruschi M, et al. Expert Rev Proteomics. 2015 Aug;12(4):425-32. doi: 10.1586/14789450.2015.1055324. Expert Rev Proteomics. 2015. PMID: 26186710 Review.
Cited by
- Carnitine palmitoyl transferase 1A is a novel diagnostic and predictive biomarker for breast cancer.
Tan Z, Zou Y, Zhu M, Luo Z, Wu T, Zheng C, Xie A, Wang H, Fang S, Liu S, Li Y, Lu Z. Tan Z, et al. BMC Cancer. 2021 Apr 15;21(1):409. doi: 10.1186/s12885-021-08134-7. BMC Cancer. 2021. PMID: 33858374 Free PMC article. - Experimental and Biological Insights from Proteomic Analyses of Extracellular Vesicle Cargos in Normalcy and Disease.
Charest A. Charest A. Adv Biosyst. 2020 Dec;4(12):e2000069. doi: 10.1002/adbi.202000069. Epub 2020 Aug 19. Adv Biosyst. 2020. PMID: 32815324 Free PMC article. Review. - Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions.
Hong J, Dauros-Singorenko P, Whitcombe A, Payne L, Blenkiron C, Phillips A, Swift S. Hong J, et al. J Extracell Vesicles. 2019 Jun 24;8(1):1632099. doi: 10.1080/20013078.2019.1632099. eCollection 2019. J Extracell Vesicles. 2019. PMID: 31275533 Free PMC article. - Immunotherapeutic interventions of Triple Negative Breast Cancer.
Li Z, Qiu Y, Lu W, Jiang Y, Wang J. Li Z, et al. J Transl Med. 2018 May 30;16(1):147. doi: 10.1186/s12967-018-1514-7. J Transl Med. 2018. PMID: 29848327 Free PMC article. Review. - Prokaryotic microvesicles Ortholog of eukaryotic extracellular vesicles in biomedical fields.
Mobarak H, Javid F, Narmi MT, Mardi N, Sadeghsoltani F, Khanicheragh P, Narimani S, Mahdipour M, Sokullu E, Valioglu F, Rahbarghazi R. Mobarak H, et al. Cell Commun Signal. 2024 Jan 30;22(1):80. doi: 10.1186/s12964-023-01414-8. Cell Commun Signal. 2024. PMID: 38291458 Free PMC article. Review.
References
- Fevrier B; Raposo G Curr. Opin. Cell Biol 2004, 16, 415–421. - PubMed
- Baietti MF; Zhang Z; Mortier E; Melchior A; Degeest G; Geeraerts A; Ivarsson Y; Depoortere F; Coomans C; Vermeiren E; Zimmermann P; David G Nat. Cell Biol 2012, 14 (7), 677–685. - PubMed
- Ostrowski M; Carmo NB; Krumeich S; Fanget I; Raposo G; Savina A; Moita CF; Schauer K; Hume AN; Freitas RP; Goud B; Benaroch P; Hacohen N; Fukuda M; Desnos C; Seabra MC; Darchen F; Amigorena S; Moita LF; Thery C Nat. Cell Biol 2010, 12 (1), 19–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG025323/AG/NIA NIH HHS/United States
- P30CA134274/CA/NCI NIH HHS/United States
- I01 BX000169/BX/BLRD VA/United States
- R01AG25323/AG/NIA NIH HHS/United States
- P30 CA134274/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials