Activator of G-Protein Signaling 3-Induced Lysosomal Biogenesis Limits Macrophage Intracellular Bacterial Infection - PubMed (original) (raw)
. 2016 Jan 15;196(2):846-56.
doi: 10.4049/jimmunol.1501595. Epub 2015 Dec 14.
Souhaila Al-Khodor 2, Gordon Y C Cheung 3, Chong-Shan Shi 1, Lalitha Srinivasan 4, Travis J McQuiston 5, Il-Young Hwang 1, Anthony J Yeh 3, Joe B Blumer 6, Volker Briken 4, Peter R Williamson 5, Michael Otto 3, Iain D C Fraser 2, John H Kehrl 7
Affiliations
- PMID: 26667172
- PMCID: PMC4811337
- DOI: 10.4049/jimmunol.1501595
Activator of G-Protein Signaling 3-Induced Lysosomal Biogenesis Limits Macrophage Intracellular Bacterial Infection
Ali Vural et al. J Immunol. 2016.
Abstract
Many intracellular pathogens cause disease by subverting macrophage innate immune defense mechanisms. Intracellular pathogens actively avoid delivery to or directly target lysosomes, the major intracellular degradative organelle. In this article, we demonstrate that activator of G-protein signaling 3 (AGS3), an LPS-inducible protein in macrophages, affects both lysosomal biogenesis and activity. AGS3 binds the Gi family of G proteins via its G-protein regulatory (GoLoco) motif, stabilizing the Gα subunit in its GDP-bound conformation. Elevated AGS3 levels in macrophages limited the activity of the mammalian target of rapamycin pathway, a sensor of cellular nutritional status. This triggered the nuclear translocation of transcription factor EB, a known activator of lysosomal gene transcription. In contrast, AGS3-deficient macrophages had increased mammalian target of rapamycin activity, reduced transcription factor EB activity, and a lower lysosomal mass. High levels of AGS3 in macrophages enhanced their resistance to infection by Burkholderia cenocepacia J2315, Mycobacterium tuberculosis, and methicillin-resistant Staphylococcus aureus, whereas AGS3-deficient macrophages were more susceptible. We conclude that LPS priming increases AGS3 levels, which enhances lysosomal function and increases the capacity of macrophages to eliminate intracellular pathogens.
Copyright © 2016 by The American Association of Immunologists, Inc.
Conflict of interest statement
Disclosures
The authors declare no competing financial interests.
Figures
Figure 1
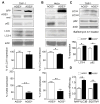
AGS3 stable cell lines exhibit increased proteolysis without an alteration in autophagic induction. (A,B,C) Immunoblot of cell lysates prepared from PMA-differentiated AGS3lo and AGS3hi THP-1 cells (A), or from AGS3lo and AGS3hi HeLa cells (B), or from Bafilomycin A1-treated AGS3lo and AGS3hi THP-1 cells (C) to examine the expression of the indicated proteins. The results obtained from three experiments are shown below and presented as % of the THP-1 AGS3lo (A) and HeLa AGS3lo (B). The LC3-II and p62 levels were normalized to the actin level in the corresponding lysates. (D) qRT-PCR of RNA prepared from AGS3lo and AGS3hi THP-1 cell lines for the indicated genes.
Figure 2
Lysosome levels are regulated by AGS3 expression. (A,B) Immunoblots of the indicated proteins in lysates from AGS3lo and AGS3hi stable THP-1 (A) and in AGS3lo and AGS3hi stable HeLa cells (B). Lysosome marker/actin band intensity ratios were normalized and the results shown as percentage of AGS3lo cells in the graphs to the right of the immunoblots. (C) Immunoblots of the indicated proteins in THP-1 stable cells expressing GFP, AGS3lo THP-1 cells, or AGS3hi THP-1 cells. (D) Confocal images of lysosomal vesicles in an AGS3lo HeLa cell (marked by asterisks) and AGS3hi (GFP positive) HeLa cell immunostained for LAMP-1 (right panel). The lysosomal mass was quantified by counting LAMP-1 positive vesicles in 50-70 cells of each cell type. The graph is shown to the right of the images. Scale bar: 10 μm. (E) Confocal images of an AGS3lo cell (** in the upper panel, visible in lower panel) and an AGS3hi THP-1 cell incubated with DQ-BSA overnight. Scale bar: 10 μm. (F) Immunoblot of indicated protein levels in cell lysates from HeLa cells transiently expressing AGS3 for 60 hours. Lysosome marker/actin band intensity ratios were normalized and shown as percentage of the control transfected cells. (G,H) Immunoblots of the indicated proteins in lysates from AGS3hi HeLa cells transfected with scrambled (ctrl) or GFP siRNAs (20nM), or HeLa cells transfected with scrambled or AGS3 siRNA (20 nM) for 72 hours. The results were normalized to actin levels and shown as a percentage of HeLa AGS3hi (G, right panel) or HeLa cells transfected with control siRNAs (H, right panel).
Figure 3
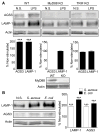
AGS3 levels alter lysosomal biogenesis by regulating the nuclear translocation of TFEB. (A) Immunoblot of the indicated proteins in cell lysates from THP-1 cells, AGS3lo and AGS3hi stable THP-1 cells; and from BMDM derived from WT and _Gpsm1_−/− mice. In some instances the BMDM were stimulated with LPS overnight (100 ng/ml). Normalized AGS3 and LAMP-1 levels are shown as a percentage of non-stimulated (left and middle) or WT (right). (B) Immunoblot of TFEB and actin expression in cell lysates prepared from the indicated cells. The LPS treated cells were from overnight stimulation. (C) qRT-PCR of RNA prepared from AGS3lo and AGS3hi THP-1 cell lines for the indicated genes. (D) Confocal images of HeLa cells expressing TFEB-GFP and AGS3-RFP or only TFEB-GFP. The location of the AGS3lo cell in the middle panel is marked by asterisks. Scale bar: 10 μm. (E) Quantification of TFEB nuclear localization in HeLa cells as in part D. Localization assessed in 500 cells. (F) Immunoblots of the indicated proteins in cytosolic and nuclear fractions isolated from HeLa cells transfected with vector or AGS3 expression plasmids overnight or treated with a GSK3β inhibitor (VIII).
Figure 4
MyD88- and TRIF-dependent signaling pathways involved in AGS3 and LAMP-1 upregulation in macrophages. (A) Representative immunoblots of cell lysates prepared from WT, MyD88, and TRIF deficient immortalized bone marrow macrophages (iBMDMs) to examine AGS3 and LAMP-1 expression. The results of three separate experiments are shown below each of the immunoblots. The data is shown as a percentage increase versus the non-stimulated cells. The cells were exposed to LPS (100 ng/ml) overnight prior to cell lysis and immunoblotting. (B) Immunoblot of cell lysates from iBMDMs exposed overnight to S. aureus, E. coli bioparticles at MOI of 2.0, or not, to examine LAMP-1 and AGS3 expression. Quantification of three separate experiments is shown to the right.
Figure 5
AGS3 regulates mTOR complex via the AKT/TSC2/mTOR pathway. (A-D) Immunoblots of the indicated proteins in cell lysates from AGS3lo and AGS3hi THP-1 cells (A), control or HeLa cells transiently expressing AGS3 for 72 hours (B), HeLa cells treated with control or siRNA targeting AGS3 expression for 72 hours (C), or BMDM from WT or _Gpsm1_−/− mice stimulated with LPS overnight (100 ng/ml). Shown below each immunoblot is the summation of results from at least 3 experiments normalized to actin and presented as a percentage of AGS3lo, % of control, % of control, or % of WT as indicated.
Figure 6
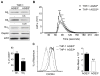
High levels of AGS3 reduce Gβ expression and inhibit chemokine receptor signaling. (A) Immunoblot of cell lysates prepared from AGS3lo and AGS3hi THP-1 cells for the expression of Gαi and Gβ. The % reduction in Gβ1 and Gβ2 expression compared to the AGS3lo cells are shown below the immunoblot. (B) Trace of the intracellular calcium levels following exposure of AGS3hi and AGS3lo cells to different concentrations of CXCL12. (C) Flow cytometry to examine CXCR4 expression on AGS3lo and AGS3hi THP-1 cells. The geometric means are shown to the right of the flow cytometry plot.
Figure 7
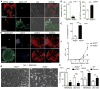
AGS3-induced lysosomal enrichment mediates resistance against antibiotic resistant bacteria. (A) Confocal microscopy images of THP-1 AGS3lo and AGS3hi cells infected with DsRed-tagged B. cenocepacia J2315 strain at MOI of 1.0 for 24 hours. The location of the AGS3lo cell is indicated with asterisks in the GFP panel. Scale bars: 10 μm. (B) The percentage of THP-1 AGS3lo and AGS3hi cells harboring ≥15 bacteria per cytosol 24 hours post-infection. (C) Quantification of CFUs from WT and _Gpsm1_−/− BMDM infected with B. cenocepacia J2315 at an MOI 1.0 at 24 hours. Results were normalized to WT cells. The results obtained from three separate infection experiments are shown for each specific bacteria. (D) Confocal microscopy images of THP-1 AGS3lo and AGS3hi cells infected with B. cenocepacia J2315 at an MOI of 1.0 at 24 hours post-infection. The cells were immunostained with B. cenocepacia and LAMP-2 specific antibodies. The location of the AGS3lo cell is indicated with asterisks in the GFP panel. Scale bars: 10 μm. (E) Quantification of the co-localization rate between B. cenocepacia J2315 and LAMP-2 (F) Kinetic analysis of CFU counts upon Mycobacterium tuberculosis infection. THP-1 AGS3lo and AGS3hi cells were infected with M. tuberculosis at MOI 0.5 for 4 hours. The cells were lysed at various time points post infection and the lysates were plated on 7H11 media for CFU counts. The results are from three independent experiments performed in triplicate. (G) Brightfield microscopy images of THP-1 AGS3lo and AGS3hi cells infected with MRSA252 at MOI of 4:1 for 6 hours. Scale bar: 400 μm. (H,I) Quantification of LDH release from THP-1 AGS3lo and AGS3hi cells (H) and from WT and G_psm1_−/− BMDM (I) 6 hours post-infection with MRSA252 or USA300 clone LAC at MOIs of 6:1 and 30:1, respectively. Control cells were incubated with a 2% Triton-X solution to determine 100% LDH release. The amounts of LDH release following bacterial infections are shown as a percentage of the Triton-X triggered release. The results obtained from three separate infection experiments are shown for each specific bacteria.
Similar articles
- Activator of G protein signaling 3 is a guanine dissociation inhibitor for Galpha i subunits.
De Vries L, Fischer T, Tronchère H, Brothers GM, Strockbine B, Siderovski DP, Farquhar MG. De Vries L, et al. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14364-9. doi: 10.1073/pnas.97.26.14364. Proc Natl Acad Sci U S A. 2000. PMID: 11121039 Free PMC article. - Normal autophagic activity in macrophages from mice lacking Gαi3, AGS3, or RGS19.
Vural A, McQuiston TJ, Blumer JB, Park C, Hwang IY, Williams-Bey Y, Shi CS, Ma DZ, Kehrl JH. Vural A, et al. PLoS One. 2013 Nov 28;8(11):e81886. doi: 10.1371/journal.pone.0081886. eCollection 2013. PLoS One. 2013. PMID: 24312373 Free PMC article. - Ric-8A catalyzes guanine nucleotide exchange on G alphai1 bound to the GPR/GoLoco exchange inhibitor AGS3.
Thomas CJ, Tall GG, Adhikari A, Sprang SR. Thomas CJ, et al. J Biol Chem. 2008 Aug 22;283(34):23150-60. doi: 10.1074/jbc.M802422200. Epub 2008 Jun 9. J Biol Chem. 2008. PMID: 18541531 Free PMC article. - Activators of G-protein signaling 3: a drug addiction molecular gateway.
Bowers MS. Bowers MS. Behav Pharmacol. 2010 Sep;21(5-6):500-13. doi: 10.1097/FBP.0b013e32833dcfa5. Behav Pharmacol. 2010. PMID: 20700046 Free PMC article. Review. - Antimicrobial responses of teleost phagocytes and innate immune evasion strategies of intracellular bacteria.
Grayfer L, Hodgkinson JW, Belosevic M. Grayfer L, et al. Dev Comp Immunol. 2014 Apr;43(2):223-42. doi: 10.1016/j.dci.2013.08.003. Epub 2013 Aug 15. Dev Comp Immunol. 2014. PMID: 23954721 Review.
Cited by
- A NOVEL NOX/PHOX-CD38-NAADP-TFEB AXIS IMPORTANT FOR MACROPHAGE ACTIVATION DURING BACTERIAL PHAGOCYTOSIS.
Najibi M, Honwad HH, Moreau JA, Becker SM, Irazoqui JE. Najibi M, et al. Autophagy. 2022 Jan;18(1):124-141. doi: 10.1080/15548627.2021.1911548. Epub 2021 Apr 13. Autophagy. 2022. PMID: 33818279 Free PMC article. - Role of G-proteins and phosphorylation in the distribution of AGS3 to cell puncta.
Vural A, Fadillioglu E, Kelesoglu F, Ma D, Lanier SM. Vural A, et al. J Cell Sci. 2018 Dec 5;131(23):jcs216507. doi: 10.1242/jcs.216507. J Cell Sci. 2018. PMID: 30404823 Free PMC article. - Knockdown of GPSM1 Inhibits the Proliferation and Promotes the Apoptosis of B-Cell Acute Lymphoblastic Leukemia Cells by Suppressing the ADCY6-RAPGEF3-JNK Signaling Pathway.
Zhang Y, Zhou B, Sun J, He Q, Zhao Y. Zhang Y, et al. Pathol Oncol Res. 2021 Apr 7;27:643376. doi: 10.3389/pore.2021.643376. eCollection 2021. Pathol Oncol Res. 2021. PMID: 34257610 Free PMC article. - The Transcription Factor EB Links Cellular Stress to the Immune Response .
Nabar NR, Kehrl JH. Nabar NR, et al. Yale J Biol Med. 2017 Jun 23;90(2):301-315. eCollection 2017 Jun. Yale J Biol Med. 2017. PMID: 28656016 Free PMC article. Review. - The impact of RGS and other G-protein regulatory proteins on Gαi-mediated signaling in immunity.
Kehrl JH. Kehrl JH. Biochem Pharmacol. 2016 Aug 15;114:40-52. doi: 10.1016/j.bcp.2016.04.005. Epub 2016 Apr 9. Biochem Pharmacol. 2016. PMID: 27071343 Free PMC article. Review.
References
- Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340:697–701. - PubMed
- Schwegmann A, Brombacher F. Host-directed drug targeting of factors hijacked by pathogens. Science signaling. 2008;1:re8. - PubMed
- Lebeis SL, Kalman D. Aligning antimicrobial drug discovery with complex and redundant host-pathogen interactions. Cell host & microbe. 2009;5:114–122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases