Golgi-Resident GTPase Rab30 Promotes the Biogenesis of Pathogen-Containing Autophagosomes - PubMed (original) (raw)
Golgi-Resident GTPase Rab30 Promotes the Biogenesis of Pathogen-Containing Autophagosomes
Seiichiro Oda et al. PLoS One. 2016.
Abstract
Autophagy acts as a host-defense system against pathogenic microorganisms such as Group A Streptococcus (GAS). Autophagy is a membrane-mediated degradation system that is regulated by intracellular membrane trafficking regulators, including small GTPase Rab proteins. Here, we identified Rab30 as a novel regulator of GAS-containing autophagosome-like vacuoles (GcAVs). We found that Rab30, a Golgi-resident Rab, was recruited to GcAVs in response to autophagy induction by GAS infection in epithelial cells. Rab30 recruitment was dependent upon its GTPase activity. In addition, the knockdown of Rab30 expression significantly reduced GcAV formation efficiency and impaired intracellular GAS degradation. Rab30 normally functions to maintain the structural integrity of the Golgi complex, but GcAV formation occurred even when the Golgi apparatus was disrupted. Although Rab30 also colocalized with a starvation-induced autophagosome, Rab30 was not required for autophagosome formation during starvation. These results suggest that Rab30 mediates autophagy against GAS independently of its normal cellular role in the structural maintenance of the Golgi apparatus, and autophagosome biogenesis during bacterial infection involves specific Rab GTPases.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
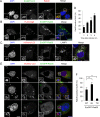
Fig 1. Golgi-resident GTPase Rab30 is localized to GAS-targeting autophagic structures.
(A) HeLa cell transiently expressing EmGFP-LC3 were infected with GAS (MOI = 100) for 4 h. Cells were then fixed, permeabilized, and immunostained with an anti-Rab30 antibody. Cellular and bacterial DNA was stained with DAPI. (B) HeLa cells transiently expressing FLAG-Atg5 and EmGFP-Rab30 were infected with GAS (MOI = 100) for 2 h. Cells were then immunostained with an anti-FLAG antibody. (C) HeLa cells transiently expressing mCherry-LC3 and EmGFP-Rab30 were infected with GAS for 4 h. Cells were then immunostained with an anti-LAMP1 antibody. Bars, 10 μm. (D) The number of cells containing GcAVs were counted and presented as the percentage of the total number of GAS-infected cells. Cells were transfected and infected with GAS for 4 h, as described in (C). The data shown represent result from >200 infected cells in terms of the mean value ± SD from 3 independent experiments. (E) HeLa cells transiently expressing mCherry-LC3 and EmGFP-Rab30 WT, EmGFP-Rab30 Q68L (QL), or EmGFP-Rab30 T23N (TN) were infected with GAS for 4 h. Bars, 10 μm. (F) Colocalization frequencies of GcAV and Rab30 were counted and presented as the percentage of the total number of GcAVs. The data shown represent the result from >80 GcAVs in terms of the mean value ± SD from 3 independent experiments. ** P < 0.01.
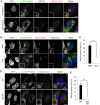
Fig 2. Rab30 is redistributed from the Golgi apparatus to GcAVs upon autophagy induction by GAS.
(A) HeLa cells transiently expressing mCherry-LC3 and EmGFP-Rab30 were infected with GAS (MOI = 100) for 4 h. Cells were immunostained with an anti-GM130 antibody. (B) HeLa cells transiently expressing EmGFP-Rab30, mCherry-Rab7, and FLAG-LC3 were infected with JRS4 WT or JRS4ΔSLO for 4 h. (C) The percentages of cells with Rab30-associated GAS were quantified. Data represent the result of >100 cells in terms of the mean value ± SD from 3 independent experiments. (D) HeLa WT or Atg5 knockout (KO) cells transiently expressing EmGFP-Rab30 and mCherry-LC3 were infected with GAS for 4 h. Bars, 10 μm. (E) The percentages of cells with Rab30-associated GAS were quantified. ** P < 0.01.
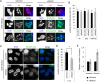
Fig 3. Rab30 is involved in autophagy during GAS infection.
(A) HeLa cells were transfected with a control siRNA or Rab30 siRNA (siRab30 #1), as well as mCherry-LC3, and EmGFP-p62, or EmGFP-NDP52 expression vector and then infected with GAS for 4 h. Cells were then fixed, permeabilized, and immunostained with an anti-ubiquitinated protein (Ub) antibody. Yellow bars, 2 μm. (B) The number of cells containing Ub/NDP52/p62-positive GAS were counted and presented as the percentage of the total number of GAS-infected cells. Data represent the results of >100 cells in terms of the mean value ± SD from 3 independent experiments. (C) HeLa cells transfected with control siRNA or Rab30 siRNA (siRab30 #1), as well as an EmGFP-LC3 expression vector were infected with GAS for 4 h. The cells were then fixed, permeabilized, and stained with DAPI. Yellow arrows indicate GcAV. Bars, 10 μm. (D) The number of cells containing GcAVs was counted and presented as the percentage of the total number of GAS. Data represent the results of >100 cells in terms of the mean value ± SD from 3 independent experiments. * P < 0.05. (E) HeLa cells were transfected with control or Rab30 siRNA (siRab30 #1). At 48 h post-transfection, HeLa cells were infected with GAS for 1, 2, or 6 h. Recovered bacteria were measured in GAS viability assays. The data shown represent the mean value ± SD from 3 independent experiments.
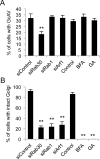
Fig 4. Involvement of Rab30 in GAS-induced autophagy is independent of its role in maintaining the structural integrity of the Golgi apparatus.
(A) HeLa cells were transfected with EmGFP-LC3 and siRNAs against the indicated target mRNAs, or were treated with BFA or GA, as indicated. Subsequently, the cells were infected with GAS (MOI = 100). The cells were then fixed, permeabilized, and stained with DAPI. The percentage of cells with GcAVs was quantified. (B) HeLa cells were transfected with siRNAs against indicated target mRNAs, or treated with BFA or GA, after which they were infected with GAS. Cells were fixed, permeabilized, and immunostained with an anti-GM130 antibody. The percentage of cells with intact Golgi structure was quantified. The data shown represent the results of >200 cells in terms of the mean value ± SD from 3 independent experiments. ** P < 0.01.
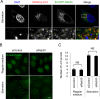
Fig 5. Involvement of Rab30 in starvation-induced autophagosome formation.
(A) Confocal microscopic images of mCherry–LC3 puncta with EmGFP–Rab30 in starvation conditions. HeLa cells that expressed mCherry–LC3 and EmGFP–Rab30 were cultured in starvation medium for 2 h and fixed. Yellow arrows indicate Rab30-colocalized LC3 puncta. (B) Confocal microscopic images of EmGFP–LC3 puncta in Rab30 knockdown cells. HeLa cells stably expressing EmGFP–LC3 were transfected with a control siRNA or Rab30 siRNA and cultured in regular medium or starvation medium for 2 h and fixed. Bars, 10 μm. (C) Effect of knockdown of the Rab30 on starvation-induced autophagosome formation. The number of LC3 dots was quantified in confocal microscopic images. The data shown represent the results for >10 images and each percentage represents the mean value ± SD based on three independent experiments. NS, not significant.
Similar articles
- RAB30 regulates PI4KB (phosphatidylinositol 4-kinase beta)-dependent autophagy against group A Streptococcus.
Nakajima K, Nozawa T, Minowa-Nozawa A, Toh H, Yamada S, Aikawa C, Nakagawa I. Nakajima K, et al. Autophagy. 2019 Mar;15(3):466-477. doi: 10.1080/15548627.2018.1532260. Epub 2018 Oct 18. Autophagy. 2019. PMID: 30290718 Free PMC article. - The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection.
Nozawa T, Aikawa C, Goda A, Maruyama F, Hamada S, Nakagawa I. Nozawa T, et al. Cell Microbiol. 2012 Aug;14(8):1149-65. doi: 10.1111/j.1462-5822.2012.01792.x. Epub 2012 Apr 17. Cell Microbiol. 2012. PMID: 22452336 - Rab17-mediated recycling endosomes contribute to autophagosome formation in response to Group A Streptococcus invasion.
Haobam B, Nozawa T, Minowa-Nozawa A, Tanaka M, Oda S, Watanabe T, Aikawa C, Maruyama F, Nakagawa I. Haobam B, et al. Cell Microbiol. 2014 Dec;16(12):1806-21. doi: 10.1111/cmi.12329. Epub 2014 Aug 30. Cell Microbiol. 2014. PMID: 25052408 - [Selective autophagy mechanism against Group A Streptococcus infection].
Nozawa T. Nozawa T. Nihon Saikingaku Zasshi. 2018;73(3):193-199. doi: 10.3412/jsb.73.193. Nihon Saikingaku Zasshi. 2018. PMID: 30158393 Review. Japanese. - Rab GTPase function in Golgi trafficking.
Barr FA. Barr FA. Semin Cell Dev Biol. 2009 Sep;20(7):780-3. doi: 10.1016/j.semcdb.2009.03.007. Semin Cell Dev Biol. 2009. PMID: 19508857 Review.
Cited by
- Rab41-mediated ESCRT machinery repairs membrane rupture by a bacterial toxin in xenophagy.
Nozawa T, Toh H, Iibushi J, Kogai K, Minowa-Nozawa A, Satoh J, Ito S, Murase K, Nakagawa I. Nozawa T, et al. Nat Commun. 2023 Oct 6;14(1):6230. doi: 10.1038/s41467-023-42039-2. Nat Commun. 2023. PMID: 37802980 Free PMC article. - NLRX1 Negatively Regulates Group A Streptococcus Invasion and Autophagy Induction by Interacting With the Beclin 1-UVRAG Complex.
Aikawa C, Nakajima S, Karimine M, Nozawa T, Minowa-Nozawa A, Toh H, Yamada S, Nakagawa I. Aikawa C, et al. Front Cell Infect Microbiol. 2018 Nov 14;8:403. doi: 10.3389/fcimb.2018.00403. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30488027 Free PMC article. - Group A Streptococcus modulates RAB1- and PIK3C3 complex-dependent autophagy.
Toh H, Nozawa T, Minowa-Nozawa A, Hikichi M, Nakajima S, Aikawa C, Nakagawa I. Toh H, et al. Autophagy. 2020 Feb;16(2):334-346. doi: 10.1080/15548627.2019.1628539. Epub 2019 Jun 14. Autophagy. 2020. PMID: 31177902 Free PMC article. - Intracellular nanovesicles mediate α5β1 integrin trafficking during cell migration.
Larocque G, Moore DJ, Sittewelle M, Kuey C, Hetmanski JHR, La-Borde PJ, Wilson BJ, Clarke NI, Caswell PT, Royle SJ. Larocque G, et al. J Cell Biol. 2021 Oct 4;220(10):e202009028. doi: 10.1083/jcb.202009028. Epub 2021 Jul 21. J Cell Biol. 2021. PMID: 34287617 Free PMC article. - Rab23's genetic structure, function and related diseases: a review.
Zheng LQ, Chi SM, Li CX. Zheng LQ, et al. Biosci Rep. 2017 Mar 2;37(2):BSR20160410. doi: 10.1042/BSR20160410. Print 2017 Apr 30. Biosci Rep. 2017. PMID: 28104793 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by the Funding Programs for Next Generation World-Leading Researchers (LS041) and JSPS KAKENHI Grant Numbers 25293370, 70294113 (to I.N.) and 10598858 (to T.N.).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials