Proteomic analysis of native cerebellar iFGF14 complexes - PubMed (original) (raw)
Proteomic analysis of native cerebellar iFGF14 complexes
Marie K Bosch et al. Channels (Austin). 2016.
Abstract
Intracellular Fibroblast Growth Factor 14 (iFGF14) and the other intracellular FGFs (iFGF11-13) regulate the properties and densities of voltage-gated neuronal and cardiac Na(+) (Nav) channels. Recent studies have demonstrated that the iFGFs can also regulate native voltage-gated Ca(2+) (Cav) channels. In the present study, a mass spectrometry (MS)-based proteomic approach was used to identify the components of native cerebellar iFGF14 complexes. Using an anti-iFGF14 antibody, native iFGF14 complexes were immunoprecipitated from wild type adult mouse cerebellum. Parallel control experiments were performed on cerebellar proteins isolated from mice (Fgf14(-/-)) harboring a targeted disruption of the Fgf14 locus. MS analyses of immunoprecipitated proteins demonstrated that the vast majority of proteins identified in native cerebellar iFGF14 complexes are Nav channel pore-forming (α) subunits or proteins previously reported to interact with Nav α subunits. In contrast, no Cav channel α or accessory subunits were revealed in cerebellar iFGF14 immunoprecipitates. Additional experiments were completed using an anti-PanNav antibody to immunoprecipitate Nav channel complexes from wild type and Fgf14(-/-) mouse cerebellum. Western blot and MS analyses revealed that the loss of iFGF14 does not measurably affect the protein composition or the relative abundance of Nav channel interacting proteins in native adult mouse cerebellar Nav channel complexes.
Keywords: cerebellum; intracellular fibroblast growth factors; native interactomes; proteomics; voltage-gated Na+ channels.
Figures
Figure 1.
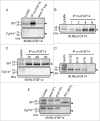
Optimization of mαFGF14 Immunoprecipitations. All blots were probed (IB) with a RbαFGF14 polyclonal antiserum as described in Materials and Methods. A. Representative Western blots of WT (top) and Fgf14 −/− (bottom) cerebellar lysates, proteins immunoprecipitated with the mouse monoclonal anti-iFGF14 (mαFGF14) antibody, and the corresponding post immunoprecipitation supernatants (post IP sup). The ∼20 kDa iFGF14 protein is clearly evident in the WT lanes and absent in the Fgf14 −/− lanes. Analyses of these blots revealed approximately 60% depletion of iFGF14 from WT mouse cerebellar lysates following IP with the mαFGF14 antibody. B. Western blots of WT cerebellar lysates before and after IP using variable amounts (1, 2, 4, or 8 mg) of protein lysates and a constant amount (20 μl) of mαFGF14-coupled sepharose beads. Analysis of IP samples revealed that increasing amounts of iFGF14 were immunoprecipitated from increasing amounts of cerebellar lysate, suggesting that the binding capacity of the mαFGF14 antibody-conjugated beads was not saturated. C. Western blots of WT (upper) and Fgf14 −/− (lower) cerebellar lysates before and after IPs using 8 mg of cerebellar proteins with variable volumes (10, 20, or 30 μl) of mαFGF14 antibody-conjugated beads. Analysis of the IP samples revealed no significant increase in the amount of iFGF14 immunoprecipitated when the bead volume was increased. D. Western blot of WT cerebellar lysates and proteins immunoprecipitated from 8 mg of cerebellar proteins with decreasing volumes of mαFGF14 antibody-conjugated beads. Non-conjugated control sepharose beads were used to maintain the bead amount constant; the numbers above the lanes refer to the mαFGF14 antibody-conjugated bead volumes (left) and the control bead volumes (right). Decreasing amounts of iFGF14 were immunoprecipitated as the mαFGF14-antibody-conjugated bead volume was decreased. E. Western blots of WT and Fgf14 −/− cerebellar lysates, post IP supernatants following sequential mαFGF14-IPs (post IP1 and post IP2), and proteins immunoprecipitated after IP2; 60% of the IP2 fraction was loaded onto the gel. Analysis of these blots revealed that approximately 85% depletion of iFGF14 was achieved with the first IP and a 90% depletion was achieved with the second IP.
Figure 2.
Mass Spectrometric Identification of iFGF14 Using In-Solution 2D-LC-MS/MS. A. Representative MS2 fragmentation spectrum of one of the identified iFGF14 tryptic peptides, NH2-(K)TKPAAHFLPKPLEVAMYR(E), with the y- (in blue) and b- (in red) ions highlighted. B. Sequence alignment of mouse iFGF11A, iFGF12A, iFGF13A, iFGF14A and iFGF14B with the amino acid sequence coverage for iFGF14 obtained following high resolution LC-MS/MS proteomic analysis of immunoprecipitated cerebellar iFGF14 complexes shown. All detected peptides are highlighted in yellow. The identified peptides that are also present in other iFGFs are also highlighted in green and the identified peptide that is unique to the iFGF14B isoform is highlighted in blue. The peptide identified by the fragmentation spectrum in A is underlined in red. The iFGF core homology domain is underlined in black. Asterisks (*) indicate fully conserved residues, colons (:) indicate residues with strongly similar properties, and periods (.) indicate residues with weakly similar properties.
Figure 3.
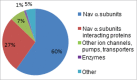
2D-LC-MS/MS Proteomic Analysis of mαFGF14-IPs. Pie chart representing the percentages of unique peptides identified in mαFGF14 antibody immunoprecipitated samples from WT cerebellum that are Nav α subunits, Nav channel accessory subunits, and other, non-Nav channel associated proteins. The vast majority (87%) of the identified peptides correspond to Nav α subunits or proteins known to associate with Nav α subunits. iFGF14 peptides are not included.
Figure 4.
Western Blot Validation of Selected mαFGF14 Immunoprecipitated Proteins Identified by High Resolution 2D-LC-MS/MS. Representative Western blots of cerebellar lysates from WT and Fgf14 −/− mice before and after IP with the mαFGF14 antibody. A. mαFGF14-IP of iFGF14 and Nav α subunits. Immunoblots (IB) with RbαFGF14 revealed a 20 kDa band (closed arrowhead) in the WT lysate and IP, but not the Fgf14 −/− lysate or IP. An additional ∼250 kDa band (open arrowhead) is present in the WT IP lane, and likely represents iFGF14 bound to Nav α subunits (see text). IB with the anti-Nav1.1, anti-Nav1.2 and anti-Nav1.6 antibodies revealed that these Nav α subunits are present at comparable levels in WT and Fgf14 −/− cerebellar lysates, but only in IP with the mαFGF14 antibody from WT cerebellum. B. IBs with the anti-Navβ1, anti-Navβ2, anti-Navβ3, anti-Navβ4 and anti-Syt2 antibodies revealed comparable levels of each of these proteins in the WT and Fgf14 −/− cerebellar lysates. Navβ1, Navβ2 and Navβ4, but not Navβ3, were also identified in Western blots following IP with the mαFGF14 antibody from WT cerebellar lysates. Two bands for Navβ2 were evident in the WT-IPs, a ∼37 kDa band (closed arrowhead) representing Navβ2 and a ∼250 kDa band (open arrowhead), likely corresponding to Navβ2 bound to Nav α subunits. IB with the anti-Syt2 antibody revealed that the ∼60 kDa Syt2 protein (closed arrowhead) was immunoprecipitated with the mαFGF14 antibody from WT and Fgf14 −/− cerebellar lysates, but was present at a higher level in the WT IP. The anti-Syt2 antibody also recognized an additional ∼250 kDa band (open arrowhead) in the WT IP, suggestive of Syt2 bound to Nav α subunits.
Figure 5.
Western Blot Analysis of Nav Channel α Subunits and Nav Channel Interacting Proteins in WT and Fgf14 −/− Cerebellar Lysates. A. Representative Western blots of cerebellar lysates from 3 WT and 3 Fgf14 −/− animals. Immunoblots (IB) with the RbαFGF14 antibody identified iFGF14 in the 3 WT lanes but not the 3 Fgf14 −/− lanes. IB with the anti-Navβ1 and anti-Navβ2 antibodies revealed no significant difference in the amounts of the Navβ1 and Navβ2 proteins in the WT and Fgf14 −/− cerebellar lysates. IB with the anti-pan-iFGF-A antibody revealed no significant difference in A-type iFGF expression in WT and Fgf14 −/− cerebellar lysates (see text). Closed arrowheads indicate dissociated protein bands and open arrowheads indicate proteins bound to Nav α subunits. B. Native Nav channel complexes were immunoprecipitated from the 3 WT and 3 Fgf14 −/− cerebellar lysates depicted in using a monoclonal anti-PanNav α subunit-specific (mαPanNav) antibody and analyzed by Western blot. Similar amounts of Nav α subunit proteins immunoprecipitate from WT and Fgf14 −/− cerebellar lysates with the mαPanNav antibody. IB with the anti-Navβ1, anti-Navβ2, anti-Navβ4 and anti-Syt2 antibodies revealed no significant differences in the amounts of the Navβ1, Navβ2, Navβ4 or Syt2 proteins that co-IP with the mαPanNav antibody from WT and Fgf14 −/− cerebellar lysates. IB with the anti-Pan-iFGF-A antibody revealed that the amount of A-type iFGFs that co-IP with the mαPanNav antibody and that remain bound to Nav α subunits (˜250 kDa; open arrowhead) are greater in the WT, compared with the Fgf14 −/−, cerebellar lysates. The gray arrowheads correspond to IgG heavy chain. C. Quantification of the intensities of the ˜250 kDa anti-Pan-iFGF-A bands, determined from blots such as those in (B), normalized to the amount of immunoprecipitated Nav α subunits in the same samples, revealed a significantly increased (*P<0.05) mean ± SEM band intensity in Fgf14 −/−, compared to WT, IPs.
Figure 6.
Optimization of mαPanNav Immunoprecipitations. A. Western blots of WT cerebellar lysates and proteins immunoprecipitated from 0.5 mg of WT cerebellar lysates with variable amounts of the mαPanNav antibody or normal mouse IgG (mIgG). Immunoblotting (IB) with the mαPanNav antibody showed no apparent increase in the amount of Nav α subunit proteins precipitating with 3 or 5 μg (compared with 1 µg) of the mαPanNav antibody. Analysis of the corresponding post IP supernatants (post IP sup) revealed that approximately 70% depletion of the Nav α subunit proteins was achieved with 3 μg of antibody. B. Western blots of WT cerebellar lysate and proteins immunoprecipitated with 3 μg of the mαPanNav antibody or mIgG from variable starting amounts of cerebellar protein. IB with the mαPanNav antibody revealed increasing amounts of Nav α subunit proteins precipitating from samples ranging from 0.25 mg to 1 mg total protein, but no further increase when the starting sample was increased to 1.5 mg protein. Analysis of the corresponding post IP supernatants (post IP sup) revealed that approximately 70% depletion of Nav α subunit proteins from the 1 mg protein sample was achieved. C. SYPRO Ruby stained gel of proteins immunoprecipitated with the mαPanNav antibody or mIgG from WT and Fgf14 −/− cerebellar lysates. Beads were eluted first with (1) 2% Rapigest, followed by (2) elution with 1% SDS. Proteins running at the molecular weight corresponding to the Nav α subunits (˜250 kDa) are clearly evident in mαPanNav-IPs from WT and Fgf14 −/− cerebella. D. Silver stained gel of proteins immunoprecipitated with the mαPanNav antibody from WT cerebellum. Precipitated proteins were analyzed from beads eluted first with 2% Rapigest followed by 1% SDS elution or beads eluted only with 2% SDS. To estimate the amount of Nav α subunit proteins in each IP sample, a bovine serum albumin (BSA) standard curve was also run. Proteins running at the molecular weight corresponding to the Nav α subunits (˜250 kDa) are clearly evident in the Rapigest and 2% SDS elutions. An estimated 50-100 ng of Nav α subunit proteins are present in the Rapigest elution.
Figure 7.
Mass Spectrometric Identification of Nav1.2 Using In-Solution 2D-LC-MS/MS. A. Representative MS2 fragmentation spectrum of one of the identified Nav1.2 tryptic peptides, corresponding to the sequence NH2-(R)SSSYHVSMDLLEDPTSR(Q) with the y- (in blue) and b- (in red) ions highlighted. B. Amino acid sequence coverage obtained for the mouse Nav1.2 protein following IP of cerebellar Nav channel complexes and LC-MS/MS proteomic analysis. Detected peptides are highlighted in yellow. The peptide for which the fragmentation spectrum is shown (in A) is underlined in red. Transmembrane segments (S1-S6) in each domain (I-IV) are underlined in black. Interdomain cytoplasmic loops I-II, II-III, and III-IV, as well as the N-terminal and carboxyl terminal domains, are also indicated.
Comment in
- iFGF14-Navs: A monogamous partnership?
Dib-Hajj SD. Dib-Hajj SD. Channels (Austin). 2016 Nov;10(6):435-6. doi: 10.1080/19336950.2016.1204829. Epub 2016 Jun 28. Channels (Austin). 2016. PMID: 27351304 Free PMC article. No abstract available.
Similar articles
- Intracellular FGF14 (iFGF14) Is Required for Spontaneous and Evoked Firing in Cerebellar Purkinje Neurons and for Motor Coordination and Balance.
Bosch MK, Carrasquillo Y, Ransdell JL, Kanakamedala A, Ornitz DM, Nerbonne JM. Bosch MK, et al. J Neurosci. 2015 Apr 29;35(17):6752-69. doi: 10.1523/JNEUROSCI.2663-14.2015. J Neurosci. 2015. PMID: 25926453 Free PMC article. - In Vivo Expression of an SCA27A-linked FGF14 Mutation Results in Haploinsufficiency and Impaired Firing of Cerebellar Purkinje Neurons.
Ransdell JL, Brown SP, Xiao M, Ornitz DM, Nerbonne JM. Ransdell JL, et al. bioRxiv [Preprint]. 2024 Oct 25:2024.10.25.620253. doi: 10.1101/2024.10.25.620253. bioRxiv. 2024. PMID: 39484407 Free PMC article. Preprint. - Mass spectrometry-based identification of native cardiac Nav1.5 channel α subunit phosphorylation sites.
Marionneau C, Lichti CF, Lindenbaum P, Charpentier F, Nerbonne JM, Townsend RR, Mérot J. Marionneau C, et al. J Proteome Res. 2012 Dec 7;11(12):5994-6007. doi: 10.1021/pr300702c. Epub 2012 Nov 9. J Proteome Res. 2012. PMID: 23092124 Free PMC article. - Proteomic analysis highlights the molecular complexities of native Kv4 channel macromolecular complexes.
Marionneau C, Townsend RR, Nerbonne JM. Marionneau C, et al. Semin Cell Dev Biol. 2011 Apr;22(2):145-52. doi: 10.1016/j.semcdb.2010.10.004. Epub 2010 Oct 17. Semin Cell Dev Biol. 2011. PMID: 20959143 Free PMC article. Review. - Current view on regulation of voltage-gated sodium channels by calcium and auxiliary proteins.
Pitt GS, Lee SY. Pitt GS, et al. Protein Sci. 2016 Sep;25(9):1573-84. doi: 10.1002/pro.2960. Epub 2016 Jun 13. Protein Sci. 2016. PMID: 27262167 Free PMC article. Review.
Cited by
- Sex-Specific Proteomic Changes Induced by Genetic Deletion of Fibroblast Growth Factor 14 (FGF14), a Regulator of Neuronal Ion Channels.
Sowers ML, Re JD, Wadsworth PA, Shavkunov AS, Lichti C, Zhang K, Laezza F. Sowers ML, et al. Proteomes. 2019 Jan 23;7(1):5. doi: 10.3390/proteomes7010005. Proteomes. 2019. PMID: 30678040 Free PMC article. - Intracellular Fibroblast Growth Factor 14: Emerging Risk Factor for Brain Disorders.
Di Re J, Wadsworth PA, Laezza F. Di Re J, et al. Front Cell Neurosci. 2017 Apr 19;11:103. doi: 10.3389/fncel.2017.00103. eCollection 2017. Front Cell Neurosci. 2017. PMID: 28469558 Free PMC article. Review. - FGF12 is a novel component of the nucleolar NOLC1/TCOF1 ribosome biogenesis complex.
Sochacka M, Karelus R, Opalinski L, Krowarsch D, Biadun M, Otlewski J, Zakrzewska M. Sochacka M, et al. Cell Commun Signal. 2022 Nov 21;20(1):182. doi: 10.1186/s12964-022-01000-4. Cell Commun Signal. 2022. PMID: 36411431 Free PMC article. - Ca2+-saturated calmodulin binds tightly to the N-terminal domain of A-type fibroblast growth factor homologous factors.
Mahling R, Rahlf CR, Hansen SC, Hayden MR, Shea MA. Mahling R, et al. J Biol Chem. 2021 Jan-Jun;296:100458. doi: 10.1016/j.jbc.2021.100458. Epub 2021 Feb 24. J Biol Chem. 2021. PMID: 33639159 Free PMC article. - NaV1.2 EFL domain allosterically enhances Ca2+ binding to sites I and II of WT and pathogenic calmodulin mutants bound to the channel CTD.
Mahling R, Hovey L, Isbell HM, Marx DC, Miller MS, Kilpatrick AM, Weaver LD, Yoder JB, Kim EH, Andresen CNJ, Li S, Shea MA. Mahling R, et al. Structure. 2021 Dec 2;29(12):1339-1356.e7. doi: 10.1016/j.str.2021.03.002. Epub 2021 Mar 25. Structure. 2021. PMID: 33770503 Free PMC article.
References
- Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev 2005; 16:215-20; PMID:15863036; http://dx.doi.org/10.1016/j.cytogfr.2005.02.002 - DOI - PMC - PubMed
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem 2003; 278:34226-36; PMID:12815063; http://dx.doi.org/10.1074/jbc.M303183200 - DOI - PubMed
- Brusse E, de Koning I, Maat-Kievit A, Oostra BA, Heutink P, van Swieten JC. Spinocerebellar ataxia associated with a mutation in the fibroblast growth factor 14 gene (SCA27): A new phenotype. Mov Disord 2006; 21:396-401; PMID:16211615; http://dx.doi.org/10.1002/mds.20708 - DOI - PubMed
- Coebergh JA, Fransen van de Putte DE, Snoeck IN, Ruivenkamp C, van Haeringen A, Smit LM. A new variable phenotype in spinocerebellar ataxia 27 (SCA 27) caused by a deletion in the FGF14 gene. Eur J Paediatr Neurol 2014; 18:413-5; PMID:24252256; http://dx.doi.org/10.1016/j.ejpn.2013.10.006 - DOI - PubMed
- Dalski A, Atici J, Kreuz FR, Hellenbroich Y, Schwinger E, Zuhlke C. Mutation analysis in the fibroblast growth factor 14 gene: frameshift mutation and polymorphisms in patients with inherited ataxias. Eur J Hum Genet 2005; 13:118-20; PMID:15470364; http://dx.doi.org/10.1038/sj.ejhg.5201286 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous