ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells - PubMed (original) (raw)
. 2016 Jun 2;12(6):936-48.
doi: 10.1080/15548627.2016.1162359. Epub 2016 May 11.
Pablo Baquero 2, Elodie M Kuntz 3, Arunima Mukhopadhyay 1, Rebecca Mitchell 2, Elaine K Allan 4, Edmond Chan 5, Kamil R Kranc 6, Bruno Calabretta 7, Paolo Salomoni 8, Eyal Gottlieb 3, Tessa L Holyoake 1, G Vignir Helgason 2
Affiliations
- PMID: 27168493
- PMCID: PMC4922442
- DOI: 10.1080/15548627.2016.1162359
ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells
Maria Karvela et al. Autophagy. 2016.
Abstract
A major drawback of tyrosine kinase inhibitor (TKI) treatment in chronic myeloid leukemia (CML) is that primitive CML cells are able to survive TKI-mediated BCR-ABL inhibition, leading to disease persistence in patients. Investigation of strategies aiming to inhibit alternative survival pathways in CML is therefore critical. We have previously shown that a nonspecific pharmacological inhibition of autophagy potentiates TKI-induced death in Philadelphia chromosome-positive cells. Here we provide further understanding of how specific and pharmacological autophagy inhibition affects nonmitochondrial and mitochondrial energy metabolism and reactive oxygen species (ROS)-mediated differentiation of CML cells and highlight ATG7 (a critical component of the LC3 conjugation system) as a potential specific therapeutic target. By combining extra- and intracellular steady state metabolite measurements by liquid chromatography-mass spectrometry with metabolic flux assays using labeled glucose and functional assays, we demonstrate that knockdown of ATG7 results in decreased glycolysis and increased flux of labeled carbons through the mitochondrial tricarboxylic acid cycle. This leads to increased oxidative phosphorylation and mitochondrial ROS accumulation. Furthermore, following ROS accumulation, CML cells, including primary CML CD34(+) progenitor cells, differentiate toward the erythroid lineage. Finally, ATG7 knockdown sensitizes CML progenitor cells to TKI-induced death, without affecting survival of normal cells, suggesting that specific inhibitors of ATG7 in combination with TKI would provide a novel therapeutic approach for CML patients exhibiting persistent disease.
Keywords: ATG7; autophagy; chronic myeloid leukemia; energy metabolism; erythroid differentiation; glycolysis; oxidative phosphorylation; reactive oxygen species; tyrosine kinase inhibitor.
Figures
Figure 1.
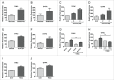
ATG7 knockdown affects extracellular metabolite levels. (A) A schematic diagram of energy metabolism in mammalian cells: During glycolysis, glucose (that contains 6 carbons) is converted into 2 molecules of the 3-carbon metabolite pyruvate via a series of intermediate metabolites. Pyruvate can then either be used to produce lactate (3-carbon molecule which is often secreted from cells) or transferred to the mitochondria for further breakdown in the TCA cycle. The TCA cycle is a series of chemical reactions, which leads to generation of energy through the oxidation of acetate (in the form of acetyl coenzyme A; acetyl Co-A) into carbon dioxide (CO2) and energy in the form of ATP, which is synthesized following OXPHOS in the mitochondrial ETC. U-13C6 can be used to examine flow of labeled carbons (indicated by red circles) through these pathways. During the conversion of 3-carbon pyruvate to acetyl Co-A, one carbon is converted to CO2 such that acetyl Co-A maintains 2 pyruvate-derived carbons (labeled) when it enters the TCA cycle. This can lead to build up of labeled carbons in TCA cycle intermediates when the pathway is activated. (B to H) sh_ATG7_-expressing K562 cells were cultured in normal medium (B to E) or in the presence of U-13C6 (F to H). Extracellular glucose (B and F), lactate (C and G), glutamate (D and H) and glutamine (E) were measured in the medium by LC-MS following 24 h culture. Note, the significance levels for metabolites with variable labeled carbons, (i.e. Glu+1 => glutamate + 1 × 13C) are indicated in figure legends to the right of the graph: +1, etc. corresponds to glutamate containing 1X, etc. 13C. Two independent experiments were performed in triplicate. P values: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Figure 2.
Autophagy inhibition affects intracellular metabolite levels. (A to G) Sh_ATG7_-expressing K562 cells were cultured in the presence of U-13C6 for 24 h. Following cell lysis, intercellular levels and incorporation of labeled carbons in glucose-6P (G6P) (A), P-enolpyruvate (PEP) (B), pyruvate (Pyr) (C), lactate (Lac) (D), citrate (Cit) (E), α-ketoglutarate (αKG) (F) and glutamate (Glu) (G) was measured by LC-MS. Two independent experiments were performed in triplicate. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. +2, etc. corresponds to metabolite containing 2X, etc. 13C.
Figure 3.
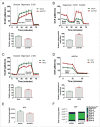
Autophagy inhibition reduces glycolysis and induces OXPHOS. ECAR (A) and OCR (B) were measured in sh_ATG7_-expressing cells using the Seahorse Extracellular Flux Analyzer. Glycolysis (relative to shCtrl-expressing cells) was calculated as “average ECAR following glucose addition minus average ECAR following inhibition of glycolysis using 2-DG.” Relative mitochondrial respiration was calculated as “basal OCR minus OCR following antimycin and rotenone (ETC inhibitors) treatment (legends for Fig. S3 provide more details). Three independent experiments were performed in quintuplicate. ECAR (C) and OCR (D) were measured in K562 cells following 24 h 10 µM HCQ treatments. Two independent experiments were performed in quintuplicate. (E and F) Sh_ATG7_-expressing K562 cells were cultured in the absence (E) or presence (F) of U-13C6 for 24 h. Following cell lysis, intercellular levels and incorporation of labeled carbons in ATP was measured by LC-MS. Two independent experiments were performed in triplicate. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. +2, etc. corresponds to ATP containing 2X, etc. 13C.
Figure 4.
Elevated mitochondrial respiration induces ROS accumulation in CML cells. (A-B) levels of active mitochondria were measured in sh_ATG7_-expressing cells following staining of cells with TMRM (A) and MTR (B) using flow cytometry. (C) Colocalization of active mitochondria and lysosomes was measured in sh_ATG7_-expressing cells following staining of cells with MTR and LAMP1 using confocal microscopy. (D to F, as well as H and I) mitochondrial superoxide levels were measured by MitoSox staining in sh_ATG7_-expressing cells (D), K562 cells following 72 h 10 µM HCQ treatment alone and in combination with 10 nM NAC (E), CP CML CD34+ cells (n = 3) following 72 h 10 µM HCQ (F), K562 cells cultured in the presence of glucose (normal medium) or galactose for 72 h (H) and K562 cells treated with 2 and 4 mM oxamate for 72 h (I). Three independent experiments (A-F, H-I) were performed in duplicate. (G) OCR was measured in K562 cells cultured in the presence of 11 mM glucose or 11 nM galactose for 72 h. Three independent experiments were performed in quintuplicate. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Figure 5.
Enhanced OXPHOS drives differentiation of K562. (A to H) cellular profile was analyzed in K562 by flow cytometry for expression of TFRC (A, C, E, G and H) and GYPA (B, D and F) following 5 d culture in galactose (A and B), 5 d treatment with 2 µM and 4 µM oxamate (C and D), ATG7 knockdown alone (E and F) or +/− 10 nM NAC treatment (G) or 72 h 10 µM HCQ treatment +/− 10 nM NAC (H). Three independent experiments were performed in duplicate. (I and J) TFRC (I) and GYPA (J) levels were measured in CP CML CD34+ cells (n = 3) following 72 h 10 µM HCQ treatment. **, P ≤ 0.01; ***, P ≤ 0.001.
Figure 6.
ATG7 knockdown enhances the effects of TKI treatment on survival of CP CML CD34+ cells. Following ATG7 knockdown CP CML CD34+ (A to C, n=3) or non-CML CD34+ cells (D, n = 3) were sorted based on GFP expression. (A) The number of autophagy-related vesicles (filled double membrane vesicles, indicated with black arrows) was quantified using electron microscopy (total of 9 cells per arm). Representative pictures are shown. (B to D) The clonogenic ability of shCtrl or sh_ATG7_ CD34+ cells was evaluated by transferring cells to semisolid media following treatment with TKI for 3 d (B, D) or 6 d (C).
Similar articles
- Altered intracellular signaling by imatinib increases the anti-cancer effects of tyrosine kinase inhibitors in chronic myelogenous leukemia cells.
Hirao T, Yamaguchi M, Kikuya M, Chibana H, Ito K, Aoki S. Hirao T, et al. Cancer Sci. 2018 Jan;109(1):121-131. doi: 10.1111/cas.13442. Epub 2017 Dec 7. Cancer Sci. 2018. PMID: 29121435 Free PMC article. - Combination of tyrosine kinase inhibitors and the MCL1 inhibitor S63845 exerts synergistic antitumorigenic effects on CML cells.
Malyukova A, Ujvari D, Yektaei-Karin E, Zovko A, Madapura HS, Keszei M, Nagy N, Lotfi K, Björn N, Wallvik J, Tamai M, Nguyen TTT, Akahane K, Inukai T, Stenke L, Salamon D. Malyukova A, et al. Cell Death Dis. 2021 Sep 25;12(10):875. doi: 10.1038/s41419-021-04154-0. Cell Death Dis. 2021. PMID: 34564697 Free PMC article. - Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells.
Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, Galavotti S, Young KW, Selmi T, Yacobi R, Van Etten RA, Donato N, Hunter A, Dinsdale D, Tirrò E, Vigneri P, Nicotera P, Dyer MJ, Holyoake T, Salomoni P, Calabretta B. Bellodi C, et al. J Clin Invest. 2009 May;119(5):1109-23. doi: 10.1172/JCI35660. Epub 2009 Apr 13. J Clin Invest. 2009. PMID: 19363292 Free PMC article. - Autophagy and mitochondrial metabolism: insights into their role and therapeutic potential in chronic myeloid leukaemia.
Baquero P, Dawson A, Helgason GV. Baquero P, et al. FEBS J. 2019 Apr;286(7):1271-1283. doi: 10.1111/febs.14659. Epub 2018 Oct 8. FEBS J. 2019. PMID: 30222247 Review. - Suppression of autophagy by BCR/ABL.
Calabretta B, Salomoni P. Calabretta B, et al. Front Biosci (Schol Ed). 2012 Jan 1;4(2):453-60. doi: 10.2741/278. Front Biosci (Schol Ed). 2012. PMID: 22202070 Free PMC article. Review.
Cited by
- Treatment-Free Remission in Chronic Myeloid Leukemia and New Approaches by Targeting Leukemia Stem Cells.
Chen Y, Zou J, Cheng F, Li W. Chen Y, et al. Front Oncol. 2021 Oct 28;11:769730. doi: 10.3389/fonc.2021.769730. eCollection 2021. Front Oncol. 2021. PMID: 34778088 Free PMC article. Review. - Targeting the Interplay between Cancer Metabolic Reprogramming and Cell Death Pathways as a Viable Therapeutic Path.
Iessi E, Vona R, Cittadini C, Matarrese P. Iessi E, et al. Biomedicines. 2021 Dec 18;9(12):1942. doi: 10.3390/biomedicines9121942. Biomedicines. 2021. PMID: 34944758 Free PMC article. Review. - The role of autophagy in cancer: from molecular mechanism to therapeutic window.
Jalali P, Shahmoradi A, Samii A, Mazloomnejad R, Hatamnejad MR, Saeed A, Namdar A, Salehi Z. Jalali P, et al. Front Immunol. 2025 Apr 3;16:1528230. doi: 10.3389/fimmu.2025.1528230. eCollection 2025. Front Immunol. 2025. PMID: 40248706 Free PMC article. Review. - A self-assembled leucine polymer sensitizes leukemic stem cells to chemotherapy by inhibiting autophagy in acute myeloid leukemia.
Xu X, Wang J, Tong T, Zhang W, Wang J, Ma W, Wang S, Zhou D, Wu J, Jiang L, Zhao M. Xu X, et al. Haematologica. 2022 Oct 1;107(10):2344-2355. doi: 10.3324/haematol.2021.280290. Haematologica. 2022. PMID: 35295079 Free PMC article. - Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients.
Mulcahy Levy JM, Thorburn A. Mulcahy Levy JM, et al. Cell Death Differ. 2020 Mar;27(3):843-857. doi: 10.1038/s41418-019-0474-7. Epub 2019 Dec 13. Cell Death Differ. 2020. PMID: 31836831 Free PMC article. Review.
References
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973; 243:290-3; PMID:4126434; http://dx.doi.org/10.1038/243290a0 - DOI - PubMed
- Konopka JB, Watanabe SM, Witte ON. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell 1984; 37:1035-42; PMID:6204766; http://dx.doi.org/10.1016/0092-8674(84)90438-0 - DOI - PubMed
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 1984; 36:93-9; PMID:6319012; http://dx.doi.org/10.1016/0092-8674(84)90077-1 - DOI - PubMed
- Holyoake TL, Helgason GV. Do we need more drugs for chronic myeloid leukemia? Immunol Rev 2015; 263:106-23; PMID:25510274; http://dx.doi.org/10.1111/imr.12234 - DOI - PubMed
- O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, Cornelissen JJ, Fischer T, Hochhaus A, Hughes T, et al.. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 2003; 348:994-1004; PMID:12637609; http://dx.doi.org/10.1056/NEJMoa022457 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0900882/MRC_/Medical Research Council/United Kingdom
- G0600782/MRC_/Medical Research Council/United Kingdom
- 18278/CRUK_/Cancer Research UK/United Kingdom
- CSO_/Chief Scientist Office/United Kingdom
- 11008/CRUK_/Cancer Research UK/United Kingdom
- 14633/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous