NCBI prokaryotic genome annotation pipeline - PubMed (original) (raw)
. 2016 Aug 19;44(14):6614-24.
doi: 10.1093/nar/gkw569. Epub 2016 Jun 24.
Affiliations
- PMID: 27342282
- PMCID: PMC5001611
- DOI: 10.1093/nar/gkw569
NCBI prokaryotic genome annotation pipeline
Tatiana Tatusova et al. Nucleic Acids Res. 2016.
Abstract
Recent technological advances have opened unprecedented opportunities for large-scale sequencing and analysis of populations of pathogenic species in disease outbreaks, as well as for large-scale diversity studies aimed at expanding our knowledge across the whole domain of prokaryotes. To meet the challenge of timely interpretation of structure, function and meaning of this vast genetic information, a comprehensive approach to automatic genome annotation is critically needed. In collaboration with Georgia Tech, NCBI has developed a new approach to genome annotation that combines alignment based methods with methods of predicting protein-coding and RNA genes and other functional elements directly from sequence. A new gene finding tool, GeneMarkS+, uses the combined evidence of protein and RNA placement by homology as an initial map of annotation to generate and modify ab initio gene predictions across the whole genome. Thus, the new NCBI's Prokaryotic Genome Annotation Pipeline (PGAP) relies more on sequence similarity when confident comparative data are available, while it relies more on statistical predictions in the absence of external evidence. The pipeline provides a framework for generation and analysis of annotation on the full breadth of prokaryotic taxonomy. For additional information on PGAP see https://www.ncbi.nlm.nih.gov/genome/annotation\_prok/ and the NCBI Handbook, https://www.ncbi.nlm.nih.gov/books/NBK174280/.
Published by Oxford University Press on behalf of Nucleic Acids Research 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures
Figure 1.
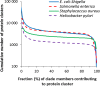
Cumulative number of protein clusters (Y) is defined for a given X (%) as the number of clusters containing proteins from fraction x ≥ X of all members of the clade. Data are presented for the four well studied clades.
Figure 2.
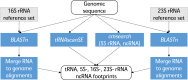
A fragment of the PGAP execution graph: prediction of structural RNA genes (ncRNA, tRNA, 5S-, 16S-, 23S- rRNA).
Figure 3.
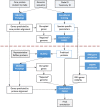
Flowchart of PGAP. The red dotted line indicates separation between pass one and pass two (see text for details).
Figure 4.
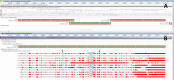
A region in the Deinococcus radiodurans R1 genome assembly (GCA_000008565.1) contains three overlapping ORFs predicted ab initio as CDSs in the first pass of PGAP. Automatic evaluation of the cross-species protein evidence through the second pass of PGAP reveals proteins bearing homology to all three fragments. Alignment of the proteins to the genome reveals otherwise unpredicted frameshifts. Green bars represent genes, red bars – coding regions; grey bars – alignments with red vertical bars indicating mismatches. (A) A region of Chromosome 1 of D. radiodurans (AE000513.1) containing the three CDS features is displayed alongside the six-frame translation. (B) The same region, updated to include final annotation markup with a frameshifted CDS as well as supporting proteins that demonstrate a consistent pattern and location of two frameshifts (marked by arrows at positions 100 733 and 100 959).
Figure 5.
Annotation of genome of Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (NC_003197). Protein alignment provides support for gene start selection. See legend to Figure 4 for description of the meaning of green, red and gray bars. (A) the first round of alignments of protein representatives from the ‘core’ protein clusters doesn't give enough evidence for gene start selection. (B) the second round of alignments clearly supports a shorter gene model which does not overlap with the upstream gene.
Figure 6.
A summary of PGAP genome annotation process is provided in the COMMENT section of GenBank and RefSeq records. The example is given for Listeria monocytogenes strain CFSAN010068, complete genome NZ_CP014250.1.
Figure 7.
Frequency histogram of genomes with respect to the fraction of the whole complement of genes supported by similarity to proteins in RefSeq. In about 50% of the total set of genomes in consideration, mostly from highly populated clades, more than 95% of protein-coding genes are supported by protein sequence similarity.
Similar articles
- RefSeq: an update on prokaryotic genome annotation and curation.
Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O'Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. Haft DH, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D851-D860. doi: 10.1093/nar/gkx1068. Nucleic Acids Res. 2018. PMID: 29112715 Free PMC article. - RefSeq and the prokaryotic genome annotation pipeline in the age of metagenomes.
Haft DH, Badretdin A, Coulouris G, DiCuccio M, Durkin AS, Jovenitti E, Li W, Mersha M, O'Neill KR, Virothaisakun J, Thibaud-Nissen F. Haft DH, et al. Nucleic Acids Res. 2024 Jan 5;52(D1):D762-D769. doi: 10.1093/nar/gkad988. Nucleic Acids Res. 2024. PMID: 37962425 Free PMC article. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - Comparison of RefSeq protein-coding regions in human and vertebrate genomes.
Fong JH, Murphy TD, Pruitt KD. Fong JH, et al. BMC Genomics. 2013 Sep 25;14:654. doi: 10.1186/1471-2164-14-654. BMC Genomics. 2013. PMID: 24063302 Free PMC article. - NCBI Taxonomy: a comprehensive update on curation, resources and tools.
Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O'Neill K, Robbertse B, Sharma S, Soussov V, Sullivan JP, Sun L, Turner S, Karsch-Mizrachi I. Schoch CL, et al. Database (Oxford). 2020 Jan 1;2020:baaa062. doi: 10.1093/database/baaa062. Database (Oxford). 2020. PMID: 32761142 Free PMC article. Review.
Cited by
- From cactus to crop: genomic insights of a beneficial and non-pathogenic Curtobacterium flaccumfaciens strain and the evolution of its pathosystem.
Ribeiro DF, de Matos JP, Rocha LCM, da Silva AK, de Paula CH, Cordeiro IF, de Carvalho Lemes CG, Sanchez AB, Garcia CCM, Setubal JC, de Souza RF, de Mello Varani A, Almeida NF, Moreira LM. Ribeiro DF, et al. Mol Genet Genomics. 2024 Nov 1;299(1):105. doi: 10.1007/s00438-024-02194-7. Mol Genet Genomics. 2024. PMID: 39485552 - Everything Is Everywhere: Physiological Responses of the Mediterranean Sea and Eastern Pacific Ocean Epiphyte Cobetia Sp. to Varying Nutrient Concentration.
Fernández-Juárez V, Jaén-Luchoro D, Brito-Echeverría J, Agawin NSR, Bennasar-Figueras A, Echeveste P. Fernández-Juárez V, et al. Microb Ecol. 2022 Feb;83(2):296-313. doi: 10.1007/s00248-021-01766-z. Epub 2021 May 5. Microb Ecol. 2022. PMID: 33954842 - Complete Genome Sequences of Three Clinical Listeria monocytogenes Sequence Type 8 Strains from Recent German Listeriosis Outbreaks.
Fischer MA, Thürmer A, Flieger A, Halbedel S. Fischer MA, et al. Microbiol Resour Announc. 2021 May 6;10(18):e00303-21. doi: 10.1128/MRA.00303-21. Microbiol Resour Announc. 2021. PMID: 33958403 Free PMC article. - Complete Genome Sequence of a Pseudomonas simiae Strain with Biocontrol Potential against Aphanomyces Root Rot.
Godebo AT, MacKenzie KD, Walley FL, Germida JJ, Yost CK. Godebo AT, et al. Microbiol Resour Announc. 2021 May 6;10(18):e00222-21. doi: 10.1128/MRA.00222-21. Microbiol Resour Announc. 2021. PMID: 33958418 Free PMC article. - MiMiC: a bioinformatic approach for generation of synthetic communities from metagenomes.
Kumar N, Hitch TCA, Haller D, Lagkouvardos I, Clavel T. Kumar N, et al. Microb Biotechnol. 2021 Jul;14(4):1757-1770. doi: 10.1111/1751-7915.13845. Epub 2021 Jun 3. Microb Biotechnol. 2021. PMID: 34081399 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources