Interactions between the Coxiella burnetii parasitophorous vacuole and the endoplasmic reticulum involve the host protein ORP1L - PubMed (original) (raw)
. 2017 Jan;19(1):10.1111/cmi.12637.
doi: 10.1111/cmi.12637. Epub 2016 Jul 15.
Affiliations
- PMID: 27345457
- PMCID: PMC5177503
- DOI: 10.1111/cmi.12637
Interactions between the Coxiella burnetii parasitophorous vacuole and the endoplasmic reticulum involve the host protein ORP1L
Anna V Justis et al. Cell Microbiol. 2017 Jan.
Abstract
Coxiella burnetii is a gram-negative intracellular bacterium that forms a large, lysosome-like parasitophorous vacuole (PV) essential for bacterial replication. Host membrane lipids are critical for the formation and maintenance of this intracellular niche, yet the mechanisms by which Coxiella manipulates host cell lipid metabolism, trafficking and signalling are unknown. Oxysterol-binding protein-related protein 1 long (ORP1L) is a mammalian lipid-binding protein that plays a dual role in cholesterol-dependent endocytic trafficking as well as interactions between endosomes and the endoplasmic reticulum (ER). We found that ORP1L localized to the Coxiella PV within 12 h of infection through a process requiring the Coxiella Dot/Icm Type 4B secretion system, which secretes effector proteins into the host cell cytoplasm where they manipulate trafficking and signalling pathways. The ORP1L N-terminal ankyrin repeats were necessary and sufficient for PV localization, indicating that ORP1L binds a PV membrane protein. Strikingly, ORP1L simultaneously co-localized with the PV and ER, and electron microscopy revealed membrane contact sites between the PV and ER membranes. In ORP1L-depleted cells, PVs were significantly smaller than PVs from control cells. These data suggest that ORP1L is specifically recruited by the bacteria to the Coxiella PV, where it influences PV membrane dynamics and interactions with the ER.
© 2016 John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures
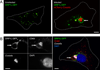
Figure 1. ORP1L localizes to the C. burnetii PV membrane
Live cell confocal (A) or fixed immunofluorescence (B) microscopy images of uninfected or infected HeLa cells expressing ORP1L-GFP. In uninfected HeLa cells, ORP1L-GFP is found on vesicular structures (A, left). ORP1L-GFP also localizes to the PV membrane (A and B, arrows) in HeLa cells infected for three days with C. burnetii expressing red fluorescent protein mCherry. Cell boundaries are shown with dotted white lines. CD63 is a marker for the PV membrane in B. Scale bars = 10 µm.
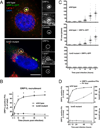
Figure 2. T4BSS-dependent recruitment of ORP1L to the PV membrane
(A) Light microscopy images of HeLa cells transfected with ORP1L-GFP and then infected with C. burnetii for 2 days. ORP1L-GFP is recruited to the PV of wild type (top) but not IcmD mutant (bottom) bacteria. Green = ORP1L-GFP, Red = Lamp1, Gray = C. burnetii, Blue = DAPI. The PV (circled) is shown magnified with individual fluorescent channels. Scale bar = 10 µm. (B) The number of ORP1L-positive PVs was quantified over a 72 hour time course of infection in HeLa cells infected with wild type (solid) or IcmD mutant (dashed) C. burnetii. Cells were transfected with ORP1L-GFP, infected, stained by immunofluorescence for C. burnetii and Lamp1, and visually scored for the presence or absence of ORP1L on the PV. Each data point represents the average of 3–4 experiments, with 20 PVs counted per experiment. Error bars represent standard error of the mean (SEM). Means compared by unpaired Welch’s t-test. * = p <0.01. (C) Measurements of PV size shows that PVs harboring wild type bacteria expand between 24 and 48 hours, while IcmD mutant PVs do not expand. HeLa cells, untransfected or expressing ORP1L-GFP, were infected with either wild type C. burnetii or the IcmD mutant. At the indicated times, the cells were fixed, stained for C. burnetii and CD63, and the PVs measured using ImageJ. PVs harboring wild type bacteria expanded between 24 and 48 hours, while IcmD PVs did not expand. Shown are individual PV measurements from three separate experiments, with at least 20 PVs per timepoint per experiment. Bars represent mean ± SEM. (D) In the comparison of ORP1L localization compared to PV expansion, ORP1L localizes to the PV 12 hours prior to significant expansion of wild type PVs (top). Error bars represent SEM.
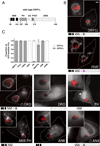
Figure 3. The ankryin repeat domains are necessary and sufficient to target ORP1L to the PV
(A) Stick diagram of ORP1L protein domains. ANK = ankryin repeats (black), PH = plekstrin homology domain (stripe), CC = coiled-coil domain (white), FFAT = two phenylalanines in an acidic tract (gray), ORD = OSBP-related ligand binding domain (dots). Numbers represents amino acid position. (B) HeLa cells were transfected with C-terminal GFP constructs and infected with mCherry-expressing C. burnetii. Domain constructs are represented below each image. At 3 days post infection, live cells were imaged by wide field fluorescence microscopy. FFAT mutant (D478A) (magenta), Ank-PH, and Ank localize to the PV membrane, while ORD, Δ Ank, and PH remain cytoplasmic without PV membrane localization. Green = ORP1L-GFP, Red = mCherry-C. burnetii. Scale bar = 10 µm. (C) Quantitation of PV localization of ORP1L domain constructs. Shown are the results from two independent experiments, with at least 30 PVs per condition per experiment. Error bars represent SEM.
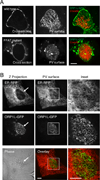
Figure 4. ORP1L simultaneously co-localizes with the PV and ER
(A) Live cell confocal microscopy images of ORP1L-GFP on the C. burnetii PV. HeLa cells were transfected with wild type ORP1L-GFP or an FFAT mutant, ORP1L(D478A)-GFP. Three days after infection with mCherry-expressing C. burnetii, the PVs were identified by phase microscopy and fluorescence imaged by live cell spinning disk confocal microscopy. When the top surface of the PV is examined, wild type ORP1L exhibits a striated pattern. This pattern is disrupted by the FFAT D478A mutation, which prevents binding to the ER protein VAP. (B) Live cell fluorescence microscopy images of _C. burnetii_-infected HeLa cells expressing ER-localized red fluorescent protein (KDEL-RFP) and ORP1L-GFP. The maximum Z projection (Z projection) shows the flattened confocal stack through the entire cell, while the PV surface is a confocal slice of the top surface of the PV (arrow). As shown in the magnification of the boxed PV, ORP1L-GFP on the PV co-localized with the host ER. Red = ER, Green = ORP1L-GFP. Scale bars = 10 µm.
Figure 5. Electron microscopy reveals membrane contact sites between the PV and ER
Transmission electron micrographs of HeLa cells infected with C. burnetii for 2 days. The boxed areas are further magnified, showing several areas where the PV membrane (outlined in blue) and ER membrane (red) are in close proximity. Cb = C. burnetii; PV = parasitophorous vacuole. Scale bars = 200 nm.
Figure 6. Depletion of ORP1L results in smaller PVs
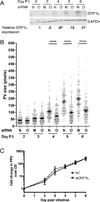
HeLa cells were treated with siRNA and then infected 2 days later (day 0 post infection (p.i.)). Infected cells were re-transfected with siRNA at 0 days p.i., and samples processed for immunoblotting, immunofluorescence, or growth assays. (A) ORP1L protein expression in cells treated with either non-targeting (N) or ORP1L siRNA (O) over a six day C. burnetii infection. Cell lysates were immunoblotted and ORP1L protein levels quantitated by normalizing to the loading control GAPDH. ORP1L protein levels remained less than 30% of the non-targeting control for the duration of the experiment. Shown is a representative blot from 6 experiments. (B) PV measurements in cells with either wild type or depleted ORP1L. At various times post infection, coverslips were fixed with paraformaldehyde and the PV membrane stained with anti-CD63 and anti-C. burnetii. The PV size was determined using ImageJ and standard error of the mean determined by ordinary one-way ANOVA with Sidak’s multiple comparisons test. The scatter plot shows individual PV measurements from three independent experiments, with at least 30 PVs per condition in each experiment. Bars indicate average ± SEM. **** = p<0.0001. (C) C. burnetii growth in ORP1L-depleted cells is similar to control cells. The number of viable bacteria was determined by fluorescent foci unit (FFU) assay at the days indicated, and normalized to day 0 to determine fold change in bacterial growth. The results are expressed as the mean of three experiments done in duplicate. Error bars represent SEM.
Figure 7. Model for ORP1L in membrane contact sites between the C. burnetii parasitophorous vacuole (PV) and host cell endoplasmic reticulum
ORP1L is recruited directly or indirectly to the C. burnetii PV by the T4BSS, where it is involved in membrane contact sites between the PV and host ER.
Similar articles
- Host Lipid Transport Protein ORP1 Is Necessary for Coxiella burnetii Growth and Vacuole Expansion in Macrophages.
Schuler B, Sladek M, Gilk SD. Schuler B, et al. mSphere. 2023 Jun 22;8(3):e0010423. doi: 10.1128/msphere.00104-23. Epub 2023 Apr 5. mSphere. 2023. PMID: 37017523 Free PMC article. - Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication.
Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. Larson CL, et al. Infect Immun. 2015 Feb;83(2):661-70. doi: 10.1128/IAI.02763-14. Epub 2014 Nov 24. Infect Immun. 2015. PMID: 25422265 Free PMC article. - Elevated Cholesterol in the Coxiella burnetii Intracellular Niche Is Bacteriolytic.
Mulye M, Samanta D, Winfree S, Heinzen RA, Gilk SD. Mulye M, et al. mBio. 2017 Feb 28;8(1):e02313-16. doi: 10.1128/mBio.02313-16. mBio. 2017. PMID: 28246364 Free PMC article. - The Coxiella burnetii parasitophorous vacuole.
Ghigo E, Colombo MI, Heinzen RA. Ghigo E, et al. Adv Exp Med Biol. 2012;984:141-69. doi: 10.1007/978-94-007-4315-1_8. Adv Exp Med Biol. 2012. PMID: 22711631 Review. - Role of lipids in Coxiella burnetii infection.
Gilk SD. Gilk SD. Adv Exp Med Biol. 2012;984:199-213. doi: 10.1007/978-94-007-4315-1_10. Adv Exp Med Biol. 2012. PMID: 22711633 Review.
Cited by
- Pathogen vacuole membrane contact sites - close encounters of the fifth kind.
Vormittag S, Ende RJ, Derré I, Hilbi H. Vormittag S, et al. Microlife. 2023 Apr 7;4:uqad018. doi: 10.1093/femsml/uqad018. eCollection 2023. Microlife. 2023. PMID: 37223745 Free PMC article. Review. - Quantitative Dextran Trafficking to the Coxiella burnetii Parasitophorous Vacuole.
Winfree S, Gilk SD. Winfree S, et al. Curr Protoc Microbiol. 2017 Aug 11;46:6C.2.1-6C.2.12. doi: 10.1002/cpmc.34. Curr Protoc Microbiol. 2017. PMID: 28800156 Free PMC article. - Organelles are miscommunicating: Membrane contact sites getting hijacked by pathogens.
Paul P, Tiwari B. Paul P, et al. Virulence. 2023 Dec;14(1):2265095. doi: 10.1080/21505594.2023.2265095. Epub 2023 Oct 20. Virulence. 2023. PMID: 37862470 Free PMC article. Review. - Phosphoregulation accommodates Type III secretion and assembly of a tether of ER-Chlamydia inclusion membrane contact sites.
Ende RJ, Murray RL, D'Spain SK, Coppens I, Derré I. Ende RJ, et al. Elife. 2022 Jul 15;11:e74535. doi: 10.7554/eLife.74535. Elife. 2022. PMID: 35838228 Free PMC article. - Hostile Takeover: Hijacking of Endoplasmic Reticulum Function by T4SS and T3SS Effectors Creates a Niche for Intracellular Pathogens.
Tsai AY, English BC, Tsolis RM. Tsai AY, et al. Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.psib-0027-2019. doi: 10.1128/microbiolspec.PSIB-0027-2019. Microbiol Spectr. 2019. PMID: 31198132 Free PMC article.
References
- Campoy EM, Mansilla ME, Colombo MI. Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol. 2013;15:922–941. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous