Heat Shock Protein 70 Family Members Interact with Crimean-Congo Hemorrhagic Fever Virus and Hazara Virus Nucleocapsid Proteins and Perform a Functional Role in the Nairovirus Replication Cycle - PubMed (original) (raw)
Heat Shock Protein 70 Family Members Interact with Crimean-Congo Hemorrhagic Fever Virus and Hazara Virus Nucleocapsid Proteins and Perform a Functional Role in the Nairovirus Replication Cycle
Rebecca Surtees et al. J Virol. 2016.
Abstract
The Nairovirus genus of the Bunyaviridae family contains serious human and animal pathogens classified within multiple serogroups and species. Of these serogroups, the Crimean-Congo hemorrhagic fever virus (CCHFV) serogroup comprises sole members CCHFV and Hazara virus (HAZV). CCHFV is an emerging zoonotic virus that causes often-fatal hemorrhagic fever in infected humans for which preventative or therapeutic strategies are not available. In contrast, HAZV is nonpathogenic to humans and thus represents an excellent model to study aspects of CCHFV biology under conditions of more-accessible biological containment. The three RNA segments that form the nairovirus genome are encapsidated by the viral nucleocapsid protein (N) to form ribonucleoprotein (RNP) complexes that are substrates for RNA synthesis and packaging into virus particles. We used quantitative proteomics to identify cellular interaction partners of CCHFV N and identified robust interactions with cellular chaperones. These interactions were validated using immunological methods, and the specific interaction between native CCHFV N and cellular chaperones of the HSP70 family was confirmed during live CCHFV infection. Using infectious HAZV, we showed for the first time that the nairovirus N-HSP70 association was maintained within both infected cells and virus particles, where N is assembled as RNPs. Reduction of active HSP70 levels in cells by the use of small-molecule inhibitors significantly reduced HAZV titers, and a model for chaperone function in the context of high genetic variability is proposed. These results suggest that chaperones of the HSP70 family are required for nairovirus replication and thus represent a genetically stable cellular therapeutic target for preventing nairovirus-mediated disease.
Importance: Nairoviruses compose a group of human and animal viruses that are transmitted by ticks and associated with serious or fatal disease. One member is Crimean-Congo hemorrhagic fever virus (CCHFV), which is responsible for fatal human disease and is recognized as an emerging threat within Europe in response to climate change. No preventative or therapeutic strategies against nairovirus-mediated disease are currently available. Here we show that the N protein of CCHFV and the related Hazara virus interact with a cellular protein, HSP70, during both the intracellular and extracellular stages of the virus life cycle. The use of inhibitors that block HSP70 function reduces virus titers by up to 1,000-fold, suggesting that this interaction is important within the context of the nairovirus life cycle and may represent a potent target for antinairovirus therapies against which the virus cannot easily develop resistance.
Copyright © 2016 Surtees et al.
Figures
FIG 1
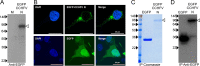
Expression and immunoprecipitation of EGFP–CCHFV-N in HEK293T cells. (A) HEK293T cells were transfected with a plasmid designed to express an EGFP–CCHFV-N fusion protein, which was detected by Western blot analysis of cell lysates using an anti-CCHFV N antibody. M, molecular mass marker lane (values at the left are in kilodaltons). (B) HEK293T cells were transfected with plasmids expressing EGFP or EGFP–CCHFV-N, and their subcellular localization was determined using confocal microscopy, with the nucleus stained blue using DAPI and with EGFP and EGFP–CCHFV-N detected as green. (C and D) Following GFP-trap-mediated IP from EGFP- and EGFP–CCHFV-N-expressing cells, proteins were resolved by SDS-PAGE and visualized by either Coomassie staining (C) or Western blotting (D) with an anti-GFP antibody. Unfilled arrowheads denote EGFP–CCHFV-N.
FIG 2
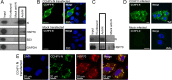
Validation of selected CCHFV N protein cellular interaction partners. HEK293T cells were transfected with EGFP–CCHFV-N and EGFP, and, 24 h later, cell lysate components were immunoprecipitated with GFP-trap, RFP-trap, or unconjugated agarose beads. The resulting IPs were separated by PAGE and then subjected to Western blotting to probe for the presence of the selected cellular proteins listed using specific antibodies. The ratios of the abundances of these proteins determined by the two SILAC LC-MS/MS analyses are indicated, and a complete list of all interaction partners is provided as an annotated table in Data set S1 in the supplemental material.
FIG 3
Interaction between cellular HSP70 and native CCHFV N expressed transiently or during CCHFV infection. (A) Anti-CCHFV N antisera was used for IP of native CCHFV N expressed in HEK293T cells, and the isolated proteins were analyzed by Western blotting using the antibodies shown. (B) The subcellular localization of native CCHFV N in HUH-7 cells was investigated using indirect immunofluorescence microscopy using anti-CCHFV N antisera, shown as green. Nuclei are stained with DAPI, shown as blue. (C) Anti-CCHFV N antisera was used for IP of native CCHFV N from CCHFV-infected SW13 cells, and the isolated proteins were analyzed by Western blotting using the antibodies shown. (D) The subcellular localization of native CCHFV N in CCHFV-infected SW13 cells was investigated by indirect immune fluorescence microscopy using anti-CCHFV N antisera, shown as green. The nuclei are stained with DAPI, shown as blue. (E) Indirect confocal immunofluorescence analysis of SW13 cells transiently expressing native CCHFV-N protein expressed from a plasmid cDNA. Cells were costained using antibodies specific for CCHFV N (green) and anti-HSP70 (red), with donkey and chicken secondary antibodies, respectively, as well as DAPI (blue).
FIG 4
Analysis of the interaction between HAZV N and HSP70 in cells. (A) HAZV N was immunoprecipitated from HAZV-infected cells using anti-N antisera and then subjected to Western blotting using the antibodies shown. (B) Indirect confocal immunofluorescence analysis of SW13 cells infected with HAZV revealed HAZV N (green) localized in discrete cytoplasmic puncta, similarly to CCHFV N. HSP70 (red) was present in both the cytoplasm and nucleus and also colocalized with HAZV N in some cytoplasmic puncta. Nuclei are stained with DAPI, shown as blue. Bottom row shows mock-infected cells.
FIG 5
SDS-PAGE analysis of purified HAZV harvested from SW13 cell supernatant and Western blot confirmation of the presence of HSP70 and HSP90 family members. (A and B) Secreted HAZV particles harvested from infected SW13 cell supernatants were purified by PEG precipitation followed by centrifugation through an iodixanol gradient. HAZV particles were isolated (lane VF1), and the remaining 4 fractions (1 ml each) were collected from the top of the gradient (Fraction 2) to the bottom of the gradient (Fraction 5). Fractions were analyzed by SDS-PAGE and either Coomassie stained (A) or silver stained (B). (C) Fraction VF1 was subjected to Western blot analysis using both anti-HAZV N antisera and antibodies targeting the listed cellular proteins, including HSP70. (D) HAZV N was immunoprecipitated from purified infectious HAZV and then subjected to Western blot analysis using anti-HAZV N and anti-HSP70 antibodies.
FIG 6
The effect of HSP70/HSC70 inhibitors VER and PIF on HAZV replication. (A) Cytotoxicity of VER and PIF in SW13 cells was assessed by MTT assay after 24 h of incubation of SW13 cells with VER and PIF at several different concentrations in the absence of HAZV. Cytotoxicity levels are presented relative to 0.4% DMSO results. (B) The effect on HAZV replication of treating A549 cells with VER and PIF was determined by plaque assay, which revealed a reduction in HAZV titers at subcytotoxic PIF and VER concentrations. The statistical significance of decreased virus multiplication using PIF and VER in relation to DMSO-only controls was assessed using paired Student's t tests, and significance is shown with an asterisk (* [P < 0.05]). (C) Western blot analysis of HAZV N protein and GAPDH levels within infected cells following PIF and VER treatment at subcytotoxic concentrations. (D) HAZV N protein levels quantified from Western blot analyses performed in three experiments.
Similar articles
- Virus-Derived DNA Forms Mediate the Persistent Infection of Tick Cells by Hazara Virus and Crimean-Congo Hemorrhagic Fever Virus.
Salvati MV, Salaris C, Monteil V, Del Vecchio C, Palù G, Parolin C, Calistri A, Bell-Sakyi L, Mirazimi A, Salata C. Salvati MV, et al. J Virol. 2021 Nov 23;95(24):e0163821. doi: 10.1128/JVI.01638-21. Epub 2021 Oct 6. J Virol. 2021. PMID: 34613808 Free PMC article. - Rescue of Infectious Recombinant Hazara Nairovirus from cDNA Reveals the Nucleocapsid Protein DQVD Caspase Cleavage Motif Performs an Essential Role other than Cleavage.
Fuller J, Surtees RA, Slack GS, Mankouri J, Hewson R, Barr JN. Fuller J, et al. J Virol. 2019 Jul 17;93(15):e00616-19. doi: 10.1128/JVI.00616-19. Print 2019 Aug 1. J Virol. 2019. PMID: 31118258 Free PMC article. - Crimean-Congo hemorrhagic fever virus nucleocapsid protein harbors distinct RNA-binding sites in the stalk and head domains.
Jeeva S, Mir S, Velasquez A, Ragan J, Leka A, Wu S, Sevarany AT, Royster AD, Almeida NA, Chan F, O'Brien L, Mir MA. Jeeva S, et al. J Biol Chem. 2019 Mar 29;294(13):5023-5037. doi: 10.1074/jbc.RA118.004976. Epub 2019 Feb 5. J Biol Chem. 2019. PMID: 30723154 Free PMC article. - Crimean-Congo Hemorrhagic Fever.
Shayan S, Bokaean M, Shahrivar MR, Chinikar S. Shayan S, et al. Lab Med. 2015 Summer;46(3):180-9. doi: 10.1309/LMN1P2FRZ7BKZSCO. Lab Med. 2015. PMID: 26199256 Review. - The Role of Nucleocapsid Protein (NP) in the Immunology of Crimean-Congo Hemorrhagic Fever Virus (CCHFV).
Pirincal A, Doymaz MZ. Pirincal A, et al. Viruses. 2024 Sep 30;16(10):1547. doi: 10.3390/v16101547. Viruses. 2024. PMID: 39459881 Free PMC article. Review.
Cited by
- Deacetylation of HSC70-4 Promotes Bombyx mori Nucleopolyhedrovirus Proliferation via Proteasome-Mediated Nuclear Import.
Mao F, Chen X, Ngowo J, Zhu Y, Lei J, Gao X, Miao M, Quan Y, Yu W. Mao F, et al. Front Physiol. 2021 Feb 19;12:609674. doi: 10.3389/fphys.2021.609674. eCollection 2021. Front Physiol. 2021. PMID: 33679433 Free PMC article. - Crimean-Congo haemorrhagic fever virus.
Hawman DW, Feldmann H. Hawman DW, et al. Nat Rev Microbiol. 2023 Jul;21(7):463-477. doi: 10.1038/s41579-023-00871-9. Epub 2023 Mar 14. Nat Rev Microbiol. 2023. PMID: 36918725 Free PMC article. Review. - A Minigenome Study of Hazara Nairovirus Genomic Promoters.
Matsumoto Y, Ohta K, Kolakofsky D, Nishio M. Matsumoto Y, et al. J Virol. 2019 Mar 5;93(6):e02118-18. doi: 10.1128/JVI.02118-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626667 Free PMC article. - Crimean-Congo Hemorrhagic Fever Virus Nucleocapsid Protein Augments mRNA Translation.
Jeeva S, Cheng E, Ganaie SS, Mir MA. Jeeva S, et al. J Virol. 2017 Jul 12;91(15):e00636-17. doi: 10.1128/JVI.00636-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28515298 Free PMC article. - Sheep and Cattle Are Not Susceptible to Experimental Inoculation with Hazara Orthonairovirus, a Tick-Borne Arbovirus Closely Related to CCHFV.
Hartlaub J, von Arnim F, Fast C, Somova M, Mirazimi A, Groschup MH, Keller M. Hartlaub J, et al. Microorganisms. 2020 Dec 4;8(12):1927. doi: 10.3390/microorganisms8121927. Microorganisms. 2020. PMID: 33291703 Free PMC article.
References
- King AM, Lefkowitz E, Adams MJ, Carstens EB. 2012. Virus taxonomy: classification and nomenclature of viruses: Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- Walker PJ, Widen SG, Wood TG, Guzman H, Tesh RB, Vasilakis N. 2016. A global genomic characterization of nairoviruses identifies nine discrete genogroups with distinctive structural characteristics and host-vector associations. Am J Trop Med Hyg 94:1107–1122. doi:10.4269/ajtmh.15-0917. - DOI - PMC - PubMed
- Suleiman MN, Muscat-Baron JM, Harries JR, Satti AG, Platt GS, Bowen ET, Simpson DI. 1980. Congo/Crimean haemorrhagic fever in Dubai. An outbreak at the Rashid Hospital. Lancet ii:939–941. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources