Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA-DNA hybrids - PubMed (original) (raw)
. 2016 Oct 14;44(18):8870-8884.
doi: 10.1093/nar/gkw752. Epub 2016 Aug 29.
Janet N Y Chan 1, Jayesh S Salvi 1, Brandon Ho 2, Amanda Hall 1, Elva Vidya 1, Ru Guo 1, Samuel A Killackey 1, Nancy Liu 1, Jeffrey E Lee 3, Grant W Brown 2, Karim Mekhail 4
Affiliations
- PMID: 27574117
- PMCID: PMC5063000
- DOI: 10.1093/nar/gkw752
Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA-DNA hybrids
Karan J Abraham et al. Nucleic Acids Res. 2016.
Abstract
Dietary calorie restriction is a broadly acting intervention that extends the lifespan of various organisms from yeast to mammals. On another front, magnesium (Mg2+) is an essential biological metal critical to fundamental cellular processes and is commonly used as both a dietary supplement and treatment for some clinical conditions. If connections exist between calorie restriction and Mg2+ is unknown. Here, we show that Mg2+, acting alone or in response to dietary calorie restriction, allows eukaryotic cells to combat genome-destabilizing and lifespan-shortening accumulations of RNA-DNA hybrids, or R-loops. In an R-loop accumulation model of Pbp1-deficient Saccharomyces cerevisiae, magnesium ions guided by cell membrane Mg2+ transporters Alr1/2 act via Mg2+-sensitive R-loop suppressors Rnh1/201 and Pif1 to restore R-loop suppression, ribosomal DNA stability and cellular lifespan. Similarly, human cells deficient in ATXN2, the human ortholog of Pbp1, exhibit nuclear R-loop accumulations repressible by Mg2+ in a process that is dependent on the TRPM7 Mg2+ transporter and the RNaseH1 R-loop suppressor. Thus, we identify Mg2+ as a biochemical signal of beneficial calorie restriction, reveal an R-loop suppressing function for human ATXN2 and propose that practical magnesium supplementation regimens can be used to combat R-loop accumulation linked to the dysfunction of disease-linked human genes.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Figure 1.
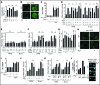
Mg2+ acting downstream or independently of calorie restriction (CR) counters R-loop accumulation. (A) CR increases intracellular Mg2+ levels and this in an Alr1/2-dependent manner. Intracellular Mg2+ levels were measured using MagnesiumGreen, a chemical that fluoresces upon binding to this cation (N = 3; Mean ± SD; _t_-test, **P < 0.01, *P < 0.05). Percentage difference relative to corresponding no Mg2+ treatment cells are indicated. (B) Confocal live cell microscopy showing that CR strongly boosts Alr1-GFP protein levels. (C) Reverse transcriptase PCR coupled to quantitative PCR (RT-qPCR) reveals that CR strongly induces Alr1 transcript levels regardless of GFP tagging (N = 3; Mean±SD). (D) Anti-RNA–DNA hybrid ChIP followed by qPCR indicates that the loss of Alr1 and Alr2 additively disrupts CR-dependent repression of R-loop buildup (N = 3; Mean ± SD; _t_-test, **P < 0.01, *P < 0.05, n.s. not statistically significant). Percentage difference relative to corresponding no Mg2+ treatment cells are indicated. (E) Loss of Alr1 and/or Alr2 does not alter R-loop levels in wild-type cells as revealed by anti-RNA–DNA hybrid ChIP (N = 3; Mean ± SD; n.s. not statistically significant for One-way ANOVA). (F) Decreasing Mg2+ concentration hinders the ability of CR to suppress RNA–DNA hybrid buildup (N = 3; Mean ± SD; _t_-test, **P < 0.01, *P < 0.05). (G) Effect of different Mg2+ supplementation regimens on intracellular Mg2+ levels as measured by Magnesium Green fluorescence (N = 3; Mean ± SD; *_t_-test P < 0.05 relative to corresponding 10 mM cells). (H) Confocal microscopy showing that 100 mM Mg2+ supplementation moderately increases Alr1-GFP protein levels. (I) RT-qPCR reveals that 100 mM Mg2+ supplementation moderately induces Alr1 transcript levels regardless of GFP tagging (N = 3; Mean ± SD). (J) Moderate Mg2+ supplementation (100 mM) partly yet significantly counters RNA–DNA hybrid accumulation in pbp1Δ cells (N = 3; Mean ± SD; *_t_-test P < 0.05). (K) A total of 100 mM Mg2+, but not 100 mM Ca2+, counters RNA–DNA hybrid accumulation in pbp1Δ cells (N = 3; Mean ± SD; *_t_-test P < 0.05). (L) A total of 100 mM Mg2+ supplementation decreases Rad52-YFP focus formation in pbp1Δ cells expressing nucleolar Nop1-CFP (N = 100; Mean ± SD).
Figure 2.
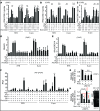
Mg2+ acts specifically via suppressors of R-loops to counter their accumulation. (A and B) Mg2+ supplementation requires Mg2+-sensitive Pif1 and Rnh1/201 to counter RNA–DNA hybrid accumulation. (C) Sir2 and Fob1 are dispensable for Mg2+-dependent suppression of RNA–DNA hybrid buildup. (D and E) Mg2+ supplementation does not alter the enrichment of RNA Pol II or diAcH3K9/14. Rnh symbolizes Rnh1/201. Control is sir2Δ. (F) Mg2+ does not decrease free long non-coding RNA (lncRNA) levels but can increase it especially in pbp1Δ sir2Δ cells. (G) Glycolytic constriction via hxk2Δ and Mg2+ supplementation redundantly suppress R-loop accumulation. (H) Mg2+ efficiently suppresses R-loops hyper-stabilized via Stm1 overexpression (left). Overexpressed Stm1 is functional as shown by rescue of cdc13-1 cell growth at 30°C (right). (A–H) Graphs show N = 3, Mean ± SD and _t_-test, **P < 0.01, *P < 0.05, n.s. not statistically significant.
Figure 3.
Mg2+ increases rDNA stability and cellular lifespan by countering R-loop buildup. (A) TAP-tagged Pif1 and Rnh1 are physically located at R-loop-accumulating rDNA loci in pbp1Δ cells under Mg2+ supplementation. R-loop-stabilizing/enriched Stm1-TAP control is included and relative fold enrichments (Mean ± SD) for RFB and E-pro are shown below gels. (B) Mg2+ counters deleterious rDNA USCE in pbp1Δ cells by operating via Pif1 and Rnh1/201. (C) Mg2+ supplementation does not decrease rDNA USCE in pbp1Δ cells lacking the R-loop stabilizer Stm1. (B and C) Shown are rates (N = 5000–25 000; Mean ± SD) and _t_-test *P < 0.05 with full data and statistics in Supplementary Table S1. Not statistically significant indicated by n.s. (D–G) Mg2+ supplementation increases the replicative lifespan of cells lacking (D) Pbp1, even in the absence of (E) Pif1 or (F) Rnh1/201, (G) but not both. (H) Combining Mg2+ supplementation with stm1Δ extends the replicative lifespan of pbp1Δ cells. (D–H) Shown are replicative lifespan curves and mean lifespans (N = 80) in parentheses with full data and Wilcoxon's rank-sum test statistics in Supplementary Table S2.
Figure 4.
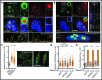
Human ATXN2 deficiency triggers nuclear R-loop accumulation. (A) Double immunofluorescence coupled to confocal microscopy reveals an inverse correlation for RNA–DNA hybrid and ATXN2 signals in HeLa cells. DAPI-stained DNA and single plane images are shown. Dashed squares outline areas shown in zoom inset images. Arrowheads highlights inverse localization of ATXN2 and R-loops even in a single nucleolus. Intensity plots below the images are for nucleolar and nucleoplasmic regions along the path of the white arrows in inset images. Bar, 5 μm. (B) Double immunofluorescence and confocal microscopy reveal that major ATXN2 foci are located within B23-outlined nucleoli of HeLa cells. DAPI-stained DNA as well as single plane, vertical slice and volume views are shown. Bar, 5 μm. (C) Short hairpin RNA (shRNA)-mediated stable knockdown of ATXN2 in HeLa cells resulted in R-loop accumulation as revealed by anti-RNA–DNA hybrid immunofluorescence and quantification of 2D images obtained by projecting confocal stacks onto a single plane. Shown are representative 2D projection images with quantification presented as a scatter plot to better capture cell population data (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, ***P < 0.001 for the Mann–Whitney test). (D) Overexpression of human RNaseH1 (+), but not empty vector control (−), counters R-loop accumulation in shATXN2 HeLa cells as revealed by anti-RNA–DNA hybrid immunofluorescence (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, ***P < 0.001, *P < 0.05 for the Mann–Whitney test). (E) Quantification analysis of nuclear signal from anti-RNaseH1 immunofluorescence indicating that overexpression of RNaseH1 (+), but not empty vector control (−), increases RNaseH1 protein levels in shCtl. and shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, *P < 0.05 for the Mann–Whitney test).
Figure 5.
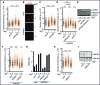
Mg2+ counters human R-loop accumulation by relying on RNaseH1 and the TRPM7 Mg2+ transporter. (A) Anti-RNA–DNA hybrid immunofluorescence indicates that 10 mM Mg2+ supplementation significantly decreases nuclear R-loop buildup in shATXN2 HeLa cells as shown in the scatter plot to better represent cell population data (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001 for the Mann–Whitney test). (B) Representative anti-ATXN2 immunofluorescence images from cells transiently transfected with siCtl. or siATXN2 in the presence or absence of 10 mM Mg2+ supplementation. Note how Mg2+ supplementation does not alter ATXN2 levels or localization. (C) Anti-γH2AX immunofluorescence imaging and quantitation indicate that 10 mM Mg2+ supplementation counters γH2AX accumulation in shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001 for the Mann–Whitney test). (D) Anti-RNA–DNA hybrid immunofluorescence reveals that transient knockdown of RNaseH1, but not PIF1, abolishes the R-loop-suppressing effect of 10 mM Mg2+ supplementation in shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, ***P < 0.001 for the Mann–Whitney test; n.s. not statistically significant). (E) Immunoblots showing transient knockdown of RNaseH1 in ATXN2-deficient HeLa cells. Anti-β-Actin immunoblotting served as loading control. (F) Anti-RNA–DNA hybrid immunofluorescence reveals that transient knockdown of TRPM7, but not MAGT1, abolishes the R-loop-suppressing effect of 10 mM Mg2+ supplementation in shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, *P < 0.05 for the Mann–Whitney test; n.s. not statistically significant). (G) RT-qPCR measuring TRPM7 RNA levels reveals that TRPM7 and MAGT1 RNA levels are unexpectedly already increased in shATXN2 HeLa cells and illustrates the efficacy of siTRPM7 in our knockdown experiments. (H) Anti-RNA–DNA hybrid immunofluorescence indicates that 10 mM Mg2+ supplementation counters nuclear R-loop buildup in siBRCA2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001 for the Mann–Whitney test). (I) Immunoblots confirming the efficacy of siRNA-mediated BRCA2 knockdown. Anti-β-Actin immunoblotting served as loading control.
Figure 6.
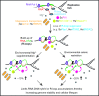
Evolutionarily conserved Mg2+-dependent process alone or in response to calorie restriction suppresses genome-destabilizing RNA–DNA hybrids, or R-loops. Loss of conserved factors such as yeast Pbp1 or human ATXN2 leads to the accumulation of genome-destabilizing RNA–DNA hybrids. Environmental calorie restriction or direct and measured Mg2+ supplementation (100 mM in yeast; 10 mM in human) increases intracellular Mg2+ levels thereby engaging Mg2+-sensitive R-loop suppressors (e.g. yeast Pif1 and Rnh1/201 or human RNaseH1) allowing them to counter R-loop accumulation and restore genome stability and cellular lifespan. Sc, S. cerevisiae; Hs, H. sapiens.
Similar articles
- Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids.
Salvi JS, Chan JN, Szafranski K, Liu TT, Wu JD, Olsen JB, Khanam N, Poon BP, Emili A, Mekhail K. Salvi JS, et al. Dev Cell. 2014 Jul 28;30(2):177-91. doi: 10.1016/j.devcel.2014.05.013. Dev Cell. 2014. PMID: 25073155 - Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1.
Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Nguyen HD, et al. Mol Cell. 2017 Mar 2;65(5):832-847.e4. doi: 10.1016/j.molcel.2017.01.029. Mol Cell. 2017. PMID: 28257700 Free PMC article. - Non-Coding RNA Molecules Connect Calorie Restriction and Lifespan.
Abraham KJ, Ostrowski LA, Mekhail K. Abraham KJ, et al. J Mol Biol. 2017 Oct 27;429(21):3196-3214. doi: 10.1016/j.jmb.2016.08.020. Epub 2016 Aug 22. J Mol Biol. 2017. PMID: 27561708 Review. - Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae.
Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Anderson RM, et al. Nature. 2003 May 8;423(6936):181-5. doi: 10.1038/nature01578. Nature. 2003. PMID: 12736687 Free PMC article. - Calorie restriction in mammals and simple model organisms.
Taormina G, Mirisola MG. Taormina G, et al. Biomed Res Int. 2014;2014:308690. doi: 10.1155/2014/308690. Epub 2014 May 6. Biomed Res Int. 2014. PMID: 24883306 Free PMC article. Review.
Cited by
- Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place.
Kotsantis P, Petermann E, Boulton SJ. Kotsantis P, et al. Cancer Discov. 2018 May;8(5):537-555. doi: 10.1158/2159-8290.CD-17-1461. Epub 2018 Apr 13. Cancer Discov. 2018. PMID: 29653955 Free PMC article. Review. - Epithelial magnesium transport by TRPM6 is essential for prenatal development and adult survival.
Chubanov V, Ferioli S, Wisnowsky A, Simmons DG, Leitzinger C, Einer C, Jonas W, Shymkiv Y, Bartsch H, Braun A, Akdogan B, Mittermeier L, Sytik L, Torben F, Jurinovic V, van der Vorst EP, Weber C, Yildirim ÖA, Sotlar K, Schürmann A, Zierler S, Zischka H, Ryazanov AG, Gudermann T. Chubanov V, et al. Elife. 2016 Dec 19;5:e20914. doi: 10.7554/eLife.20914. Elife. 2016. PMID: 27991852 Free PMC article. - The role of DNA damage response in amyotrophic lateral sclerosis.
Sun Y, Curle AJ, Haider AM, Balmus G. Sun Y, et al. Essays Biochem. 2020 Oct 26;64(5):847-861. doi: 10.1042/EBC20200002. Essays Biochem. 2020. PMID: 33078197 Free PMC article. Review. - Nucleolar RNA polymerase II drives ribosome biogenesis.
Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, Wang M, Bokros M, Vidya E, Ostrowski LA, Oshidari R, Pietrobon V, Patel PS, Algouneh A, Singhania R, Liu Y, Yerlici VT, De Carvalho DD, Ohh M, Dickson BC, Hakem R, Greenblatt JF, Lee S, Bishop AJR, Mekhail K. Abraham KJ, et al. Nature. 2020 Sep;585(7824):298-302. doi: 10.1038/s41586-020-2497-0. Epub 2020 Jul 15. Nature. 2020. PMID: 32669707 Free PMC article. - DNA damage and regulation of protein homeostasis.
Paull TT. Paull TT. DNA Repair (Amst). 2021 Sep;105:103155. doi: 10.1016/j.dnarep.2021.103155. Epub 2021 Jun 8. DNA Repair (Amst). 2021. PMID: 34116476 Free PMC article. Review.
References
- Lin S.J., Kaeberlein M., Andalis A.A., Sturtz L.A., Defossez P.A., Culotta V.C., Fink G.R., Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. - PubMed
- Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous