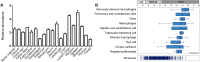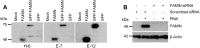Functional Analyses of the Crohn's Disease Risk Gene LACC1 - PubMed (original) (raw)
. 2016 Dec 13;11(12):e0168276.
doi: 10.1371/journal.pone.0168276. eCollection 2016.
Liselotte Vesterlund 1, Ferdinando Bonfiglio 1, Luca Mazzurana 2, Lina Cordeddu 1, Danika Schepis 3, Jenny Mjösberg 2, Sabrina Ruhrmann 4, Alessia Fabbri 5, Vladana Vukojevic 4, Piergiorgio Percipalle 6 7, Florian A Salomons 8, Jurga Laurencikiene 9, Leif Törkvist 10, Jonas Halfvarson 11, Mauro D'Amato 1 12
Affiliations
- PMID: 27959965
- PMCID: PMC5154582
- DOI: 10.1371/journal.pone.0168276
Functional Analyses of the Crohn's Disease Risk Gene LACC1
Ghazaleh Assadi et al. PLoS One. 2016.
Abstract
Background: Genetic variation in the Laccase (multicopper oxidoreductase) domain-containing 1 (LACC1) gene has been shown to affect the risk of Crohn's disease, leprosy and, more recently, ulcerative colitis and juvenile idiopathic arthritis. LACC1 function appears to promote fatty-acid oxidation, with concomitant inflammasome activation, reactive oxygen species production, and anti-bacterial responses in macrophages. We sought to contribute to elucidating LACC1 biological function by extensive characterization of its expression in human tissues and cells, and through preliminary analyses of the regulatory mechanisms driving such expression.
Methods: We implemented Western blot, quantitative real-time PCR, immunofluorescence microscopy, and flow cytometry analyses to investigate fatty acid metabolism-immune nexus (FAMIN; the LACC1 encoded protein) expression in subcellular compartments, cell lines and relevant human tissues. Gene-set enrichment analyses were performed to initially investigate modulatory mechanisms of LACC1 expression. A small-interference RNA knockdown in vitro model system was used to study the effect of FAMIN depletion on peroxisome function.
Results: FAMIN expression was detected in macrophage-differentiated THP-1 cells and several human tissues, being highest in neutrophils, monocytes/macrophages, myeloid and plasmacytoid dendritic cells among peripheral blood cells. Subcellular co-localization was exclusively confined to peroxisomes, with some additional positivity for organelle endomembrane structures. LACC1 co-expression signatures were enriched for genes involved in peroxisome proliferator-activated receptors (PPAR) signaling pathways, and PPAR ligands downregulated FAMIN expression in in vitro model systems.
Conclusion: FAMIN is a peroxisome-associated protein with primary role(s) in macrophages and other immune cells, where its metabolic functions may be modulated by PPAR signaling events. However, the precise molecular mechanisms through which FAMIN exerts its biological effects in immune cells remain to be elucidated.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Fig 1. LACC1 mRNA expression in immune-related and gastrointestinal human tissues.
(A) qRT-PCR analysis of LACC1 relative expression in a panel of human tissues identifies spleen and lymph nodes as tissues with higher LACC1 expression. Reactions were done in triplicates. (B) LACC1 mRNA expression data extracted from Genevisible (
https://genevestigator.com/gv/
).
Fig 2. Characterization of anti-FAMIN antibodies and FAMIN expression in HeLa and THP-1 cell lines.
(A) Anti-FAMIN mouse monoclonal antibodies (H-6, E-7 and E-12) were used in WB analyses of cell extracts from HeLa cells untransfected (mock) or transfected with FAMIN, GFP-FAMIN or GFP (control vector). (B) FAMIN expression is induced upon PMA differentiation of THP-1 cells (lanes 1–2 cells transfected with scrambled-control siRNA). Complete knockdown of FAMIN expression was obtained upon transfection of THP-1 cells with siRNA targeting LACC1 transcript (lanes 3–4). β-actin was used for equal loading control. Data are representative of three independent experiments.
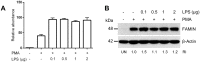
Fig 3. LPS stimulation of THP-1 cells results in FAMIN upregulation.
(A) qRT-PCR analysis of LACC1 mRNA expression in THP-1 cells stimulated with PMA and LPS. (B) WB analysis of FAMIN protein expression in THP-1 cells stimulated with PMA and LPS. Relative intensity (RI) from densitometric analysis is reported. Data are representative of three independent experiments. Abbreviation; UN: unstimulated control.
Fig 4. FACS analyses of FAMIN expression in PBMCs and granulocytes.
PBMCs and granulocytes from human healthy donors were co-stained for different cell markers and FAMIN. Neutrophils, monocytes and DCs were shown to be FAMIN+. The events in the displayed graphs were first identified and gated by forward and side scatter parameters. (A) Gating strategy for FAMIN+ monocytes (CD14+, black) and CD14- neutrophils (CD15+, purple and CD16+, orange). (B) Gating strategy for FAMIN+ myeloid DCs (CD11c+, brown) and plasmacytoid DCs (BDCA2+, green) was obtained from gating the CD3-CD19- population followed by gating of CD14-HLADR+ cells. (C) Gating strategy for FAMIN+ T-cells (CD3+, purple) and B-cells (CD19+, orange). Numbers in the outlined areas indicate percent cells in each cell subtype. Data are representative of three independent experiments.
Fig 5. Characterization of FAMIN subcellular localization.
PMA differentiated THP-1 cells were co-stained with a panel of antibodies directed towards different organelle markers and anti-FAMIN antibodies. Magnifications are shown for each staining on the right side of the pictures, together with indication of the target protein and corresponding compartment or cell organelle. Co-localization of FAMIN with PMP70 and catalase were previously shown and serve as positive control, Some additional co-localization could also be detected for endomembrane structure proteins detected at the level of endoplasmic reticulum (ER; calnexin, calreticulin and PDI), lysosomes (LAMP-1) and mitochondrion (COX-IV). Data are representative of six independent experiments.
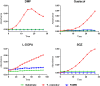
Fig 6. Analysis of human recombinant FAMIN laccase activity.
Four phenolic substrates (DMP, Guaiacol, L-DOPA and SGZ) were tested in order to evaluate laccase activity of the C-terminal MYC/DDK-tagged recombinant FAMIN protein, as indicated. Data are representative of three independent experiments.
Fig 7. Effect of PPAR agonists FAMIN expression in THP-1 cells.
WB analysis of FAMIN expression in THP-1 macrophages after 24hrs treatment with PPARα (WY14643) or PPARγ (rosiglitazone) as indicated. β-actin were used as a loading control. Relative intensity (RI) from densitometric analysis is reported. Data are representative of three independent experiments. Abbreviation; UN: unstimulated control.
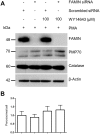
Fig 8. Effect of siRNA-mediated FAMIN knockdown on peroxisome number and peroxisome protein expression.
(A) WB analysis of PMA-induced THP-1 macrophages show diminished FAMIN expression upon siRNA knockdown. Subsequent treatment with WY14643 does not affect peroxisome protein expression. (B) However, a possible trend towards increasing numbers of peroxisomes per cell in THP-1 macrophages could be observed after treatment with PPARα agonist WY14643, independent of FAMIN presence or absence. Data are representative of three independent experiments.
Similar articles
- C13orf31 (FAMIN) is a central regulator of immunometabolic function.
Cader MZ, Boroviak K, Zhang Q, Assadi G, Kempster SL, Sewell GW, Saveljeva S, Ashcroft JW, Clare S, Mukhopadhyay S, Brown KP, Tschurtschenthaler M, Raine T, Doe B, Chilvers ER, Griffin JL, Kaneider NC, Floto RA, D'Amato M, Bradley A, Wakelam MJ, Dougan G, Kaser A. Cader MZ, et al. Nat Immunol. 2016 Sep;17(9):1046-56. doi: 10.1038/ni.3532. Epub 2016 Aug 1. Nat Immunol. 2016. PMID: 27478939 Free PMC article. - Biallelic loss-of-function LACC1/FAMIN Mutations Presenting as Rheumatoid Factor-Negative Polyarticular Juvenile Idiopathic Arthritis.
Rabionet R, Remesal A, Mensa-Vilaró A, Murías S, Alcobendas R, González-Roca E, Ruiz-Ortiz E, Antón J, Iglesias E, Modesto C, Comas D, Puig A, Drechsel O, Ossowski S, Yagüe J, Merino R, Estivill X, Arostegui JI. Rabionet R, et al. Sci Rep. 2019 Mar 14;9(1):4579. doi: 10.1038/s41598-019-40874-2. Sci Rep. 2019. PMID: 30872671 Free PMC article. - Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes.
Lahiri A, Hedl M, Yan J, Abraham C. Lahiri A, et al. Nat Commun. 2017 Jun 8;8:15614. doi: 10.1038/ncomms15614. Nat Commun. 2017. PMID: 28593945 Free PMC article. - LACC1: A critical involvement in macrophage immunometabolism.
Li Y, Wu Z, Tan X, Tang L, Ouyang F. Li Y, et al. Cell Biol Int. 2023 Sep;47(9):1488-1490. doi: 10.1002/cbin.12063. Epub 2023 Jun 27. Cell Biol Int. 2023. PMID: 37366569 Review. - Using genes to triangulate the pathophysiology of granulomatous autoinflammatory disease: NOD2, PLCG2 and LACC1.
Szymanski AM, Ombrello MJ. Szymanski AM, et al. Int Immunol. 2018 Apr 25;30(5):205-213. doi: 10.1093/intimm/dxy021. Int Immunol. 2018. PMID: 29538758 Free PMC article. Review.
Cited by
- Genetic Susceptibility to Leprosy-From Classic Immune-Related Candidate Genes to Hypothesis-Free, Whole Genome Approaches.
Cambri G, Mira MT. Cambri G, et al. Front Immunol. 2018 Jul 20;9:1674. doi: 10.3389/fimmu.2018.01674. eCollection 2018. Front Immunol. 2018. PMID: 30079069 Free PMC article. Review. - Myeloid Cell Expression of LACC1 Is Required for Bacterial Clearance and Control of Intestinal Inflammation.
Kang JW, Yan J, Ranjan K, Zhang X, Turner JR, Abraham C. Kang JW, et al. Gastroenterology. 2020 Sep;159(3):1051-1067. doi: 10.1053/j.gastro.2020.07.024. Epub 2020 Jul 18. Gastroenterology. 2020. PMID: 32693188 Free PMC article. - LACC1 Regulates TNF and IL-17 in Mouse Models of Arthritis and Inflammation.
Skon-Hegg C, Zhang J, Wu X, Sagolla M, Ota N, Wuster A, Tom J, Doran E, Ramamoorthi N, Caplazi P, Monroe J, Lee WP, Behrens TW. Skon-Hegg C, et al. J Immunol. 2019 Jan 1;202(1):183-193. doi: 10.4049/jimmunol.1800636. Epub 2018 Dec 3. J Immunol. 2019. PMID: 30510070 Free PMC article. - Laccase Properties, Physiological Functions, and Evolution.
Janusz G, Pawlik A, Świderska-Burek U, Polak J, Sulej J, Jarosz-Wilkołazka A, Paszczyński A. Janusz G, et al. Int J Mol Sci. 2020 Jan 31;21(3):966. doi: 10.3390/ijms21030966. Int J Mol Sci. 2020. PMID: 32024019 Free PMC article. Review. - Single-Nucleotide Polymorphisms Related to Leprosy Risk and Clinical Phenotypes Among Chinese Population.
Long SY, Wang L, Jiang HQ, Shi Y, Zhang WY, Xiong JS, Sun PW, Chen YQ, Mei YM, Pan C, Ge G, Wang ZZ, Wu ZW, Yu MW, Wang HS. Long SY, et al. Pharmgenomics Pers Med. 2021 Jul 12;14:813-821. doi: 10.2147/PGPM.S314861. eCollection 2021. Pharmgenomics Pers Med. 2021. PMID: 34285550 Free PMC article.
References
- Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. Elsevier Inc.; 2012;142: 46–54. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases