Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin - PubMed (original) (raw)
Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin
Hana Hanzlikova et al. Nucleic Acids Res. 2017.
Abstract
A critical step of DNA single-strand break repair is the rapid recruitment of the scaffold protein XRCC1 that interacts with, stabilizes and stimulates multiple enzymatic components of the repair process. XRCC1 recruitment is promoted by PARP1, an enzyme that is activated following DNA damage and synthesizes ADP-ribose polymers that XRCC1 binds directly. However, cells possess two other DNA strand break-induced PARP enzymes, PARP2 and PARP3, for which the roles are unclear. To address their involvement in the recruitment of endogenous XRCC1 into oxidized chromatin we have established 'isogenic' human diploid cells in which PARP1 and/or PARP2, or PARP3 are deleted. Surprisingly, we show that either PARP1 or PARP2 are sufficient for near-normal XRCC1 recruitment at oxidative single-strand breaks (SSBs) as indicated by the requirement for loss of both proteins to greatly reduce or ablate XRCC1 chromatin binding following H2O2 treatment. Similar results were observed for PNKP; an XRCC1 protein partner important for repair of oxidative SSBs. Notably, concentrations of PARP inhibitor >1000-fold higher than the IC50 were required to ablate both ADP-ribosylation and XRCC1 chromatin binding following H2O2 treatment. These results demonstrate that very low levels of ADP-ribosylation, synthesized by either PARP1 or PARP2, are sufficient for XRCC1 recruitment following oxidative stress.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
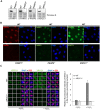
Figure 1.
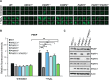
Development of PARP1-/-, PARP2-/-, PARP3-/- and XRCC1-/- RPE-1 cells and XRCC1 high-content imaging. Wild type (WT), PARP1-/-, PARP2-/-, PARP3-/- and XRCC1-/- RPE-1 clonal cell lines were analysed for loss of the targeted protein by (A) Western blotting and (B) immunofluorescence. Note that the PARP3 antibody available to us was not suitable for immunofluorescence. (C) Left, representative ScanR images of WT and XRCC1-/- RPE-1 cells non-treated or treated with 1 mM hydrogen peroxide (H2O2) for 10 min and pre-extracted with detergent prior to fixation and immunostaining for XRCC1 (green), the nucleolar marker B23 (red) and counterstaining with DAPI (blue). Right, quantification of detergent-insoluble anti-XRCC1 signal (excluding nucleolar XRCC1 signal) from >1000 cells per sample using Olympus ScanR analysis software. Data are the mean (±SEM) of three independent experiments. The black dotted line denotes non-specific anti-XRCC1 background signal, defined as the residual signal in XRCC1-/- RPE-1 cells.
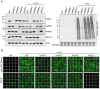
Figure 2.
Levels of H2O2-induced ADP-ribosylation and XRCC1 recruitment into chromatin in PARP-deleted RPE-1 cells. (A) Levels of the indicated proteins (left) and ADP-ribosylated proteins (right) were compared in cell lysates from the indicated WT or mutant RPE-1 cells harvested before and after treatment with 400 µM H2O2 for 7 min by Western blotting using appropriate antibodies and anti-pan-ADP-ribose binding reagent. (B) Levels of ADP-ribosylation and chromatin-bound XRCC1 were analysed by indirect immunofluorescence in cells treated or not with H2O2 (as above) by fixation and staining with anti-pan-ADP-ribose binding reagent (top panels) or by detergent pre-extraction prior to fixation and staining with anti-XRCC1 antibody (bottom panels). Representative ScanR images are shown.
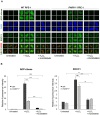
Figure 3.
Residual recruitment of endogenous XRCC1 into oxidized chromatin in PARP1-/- RPE-1 cells is greatly reduced by PARP inhibitor. (A) WT and PARP1-/- RPE-1 cells were pre-incubated or not with 10 μM KU0058948 inhibitor for 1 h prior to a 7 min incubation with or without 400 μM H2O2. Cells were pre-extracted with detergent to remove non-chromatin bound proteins prior to fixation and immunostaining with the indicated antibodies or anti-pan-ADP-ribose binding reagent. Representative ScanR images are shown. (B) Quantification of total nuclear pan-ADP-ribose and chromatin-bound nuclear XRCC1 (excluding nucleolar XRCC1 signal) in cells treated as in panel A. Nucleoli were located using anti-B23 antibodies. All data are the mean (±SEM) of three independent experiments with >1000 cells scored per sample in each experiment. Statistical significance was assessed by two-tailed t-tests. Asterisks ** and *** indicate _P_-values of <0.01 and <0.001, respectively; ns – not significant. The black dotted line denotes non-specific anti-XRCC1 background signal, defined as the residual signal in XRCC1-/- cells stained in parallel.
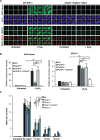
Figure 4.
Overlapping roles for PARP1 and PARP2 in recruiting endogenous XRCC1 into oxidized chromatin. (A) Levels of ADP-ribosylation and chromatin-bound XRCC1 were measured by indirect immunofluorescence in WT and _PARP1-/-/PARP2-/-_cells treated or not with 400 μM H2O2 for 7 min by fixation and staining with anti-pan-ADP-ribose binding reagent and DAPI (top panels) or by detergent pre-extraction prior to fixation and staining with anti-XRCC1 and anti-B23 antibodies (bottom panels). Representative ScanR images are shown. (B) Quantification of total nuclear pan-ADP-ribose and chromatin bound XRCC1 (excluding nucleolar signal). The black dotted line denotes non-specific anti-XRCC1 background signal, measured by XRCC1 immunostaining in XRCC1-/- cells in parallel. All data are the mean (±SEM) of three independent experiments with >1000 cells scored per sample in each experiment. Statistical significance was assessed by two tailed t-tests. Asterisks * and ** indicate _P_-values of <0.05 and <0.01, respectively; ns – not significant. (C) DNA strand breakage was quantified by alkaline comet assays in indicated RPE-1 cells before, immediately after treatment with 50 μM H2O2 on ice and after the depicted repair periods in drug-free medium. Data are the average comet tail moment (an arbitrary unit-measure of DNA strand breaks) of 100 cells per sample and are the mean (±SEM) of three independent experiments. Statistically significant differences (two-way ANOVA) are indicated (**P < 0.01; ***P < 0.001; ****P < 0.0001; ns – not significant).
Figure 5.
Overlapping roles for PARP1 and PARP2 in recruiting endogenous PNKP into oxidised chromatin. (A) Levels of chromatin-bound PNKP were analysed by indirect immunofluorescence in indicated RPE-1 cell lines untreated or treated with 400 μM H2O2 for 7 min by detergent pre-extraction prior to fixation and staining with anti-PNKP antibody. Representative ScanR images are shown. (B) Quantification of chromatin-bound PNKP in cells measured as above. All data are the mean (±SEM) of three independent experiments with >2000 cells scored per sample in each experiment. Statistical significance was assessed by two-tailed t-tests. Asterisks * and ** indicate _P_-values of <0.05 and <0.01, respectively; ns – not significant. (C) Levels of PNKP and the relevant proteins in cell extracts from RPE-1 cells of the indicated genotype.
Similar articles
- XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia.
Hoch NC, Hanzlikova H, Rulten SL, Tétreault M, Komulainen E, Ju L, Hornyak P, Zeng Z, Gittens W, Rey SA, Staras K, Mancini GM, McKinnon PJ, Wang ZQ, Wagner JD; Care4Rare Canada Consortium; Yoon G, Caldecott KW. Hoch NC, et al. Nature. 2017 Jan 5;541(7635):87-91. doi: 10.1038/nature20790. Epub 2016 Dec 21. Nature. 2017. PMID: 28002403 Free PMC article. - XRCC1-mediated repair of strand breaks independent of PNKP binding.
Horton JK, Stefanick DF, Zhao ML, Janoshazi AK, Gassman NR, Seddon HJ, Wilson SH. Horton JK, et al. DNA Repair (Amst). 2017 Dec;60:52-63. doi: 10.1016/j.dnarep.2017.10.007. Epub 2017 Oct 19. DNA Repair (Amst). 2017. PMID: 29100039 Free PMC article. - Dispensability of HPF1 for cellular removal of DNA single-strand breaks.
Hrychova K, Burdova K, Polackova Z, Giamaki D, Valtorta B, Brazina J, Krejcikova K, Kuttichova B, Caldecott KW, Hanzlikova H. Hrychova K, et al. Nucleic Acids Res. 2024 Oct 14;52(18):10986-10998. doi: 10.1093/nar/gkae708. Nucleic Acids Res. 2024. PMID: 39162207 Free PMC article. - Mechanistic insight into the role of Poly(ADP-ribosyl)ation in DNA topology modulation and response to DNA damage.
Matkarimov BT, Zharkov DO, Saparbaev MK. Matkarimov BT, et al. Mutagenesis. 2020 Feb 13;35(1):107-118. doi: 10.1093/mutage/gez045. Mutagenesis. 2020. PMID: 31782485 Review. - The dynamics and regulation of PARP1 and PARP2 in response to DNA damage and during replication.
Zhang H, Zha S. Zhang H, et al. DNA Repair (Amst). 2024 Aug;140:103690. doi: 10.1016/j.dnarep.2024.103690. Epub 2024 May 25. DNA Repair (Amst). 2024. PMID: 38823186 Review.
Cited by
- Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach.
Lodovichi S, Cervelli T, Pellicioli A, Galli A. Lodovichi S, et al. Int J Mol Sci. 2020 Sep 12;21(18):6684. doi: 10.3390/ijms21186684. Int J Mol Sci. 2020. PMID: 32932697 Free PMC article. Review. - PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance.
Rose M, Burgess JT, O'Byrne K, Richard DJ, Bolderson E. Rose M, et al. Front Cell Dev Biol. 2020 Sep 9;8:564601. doi: 10.3389/fcell.2020.564601. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33015058 Free PMC article. Review. - TET-mediated DNA hydroxymethylation is negatively influenced by the PARP-dependent PARylation.
Tolić A, Ravichandran M, Rajić J, Đorđević M, Đorđević M, Dinić S, Grdović N, Jovanović JA, Mihailović M, Nestorović N, Jurkowski TP, Uskoković AS, Vidaković MS. Tolić A, et al. Epigenetics Chromatin. 2022 Apr 5;15(1):11. doi: 10.1186/s13072-022-00445-8. Epigenetics Chromatin. 2022. PMID: 35382873 Free PMC article. - Dual function of HPF1 in the modulation of PARP1 and PARP2 activities.
Kurgina TA, Moor NA, Kutuzov MM, Naumenko KN, Ukraintsev AA, Lavrik OI. Kurgina TA, et al. Commun Biol. 2021 Nov 3;4(1):1259. doi: 10.1038/s42003-021-02780-0. Commun Biol. 2021. PMID: 34732825 Free PMC article. - The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage.
Menoni H, Wienholz F, Theil AF, Janssens RC, Lans H, Campalans A, Radicella JP, Marteijn JA, Vermeulen W. Menoni H, et al. Nucleic Acids Res. 2018 Sep 6;46(15):7747-7756. doi: 10.1093/nar/gky579. Nucleic Acids Res. 2018. PMID: 29955842 Free PMC article.
References
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. - PubMed
- Caldecott K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008; 9:619–631. - PubMed
- Caldecott K.W. Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair (Amst). 2014; 19:108–113. - PubMed
- de Murcia G., Menissier-de Murcia J.. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 1994; 19:172–176. - PubMed
- Amé J.-C., Spenlehauer C., de Murcia G.. The PARP superfamily. Bioessays. 2004; 26:882–893. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous