The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes - PubMed (original) (raw)
The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes
Rituraj Marwaha et al. J Cell Biol. 2017.
Abstract
Endocytic, autophagic, and phagocytic vesicles move on microtubule tracks to fuse with lysosomes. Small GTPases, such as Rab7 and Arl8b, recruit their downstream effectors to mediate this transport and fusion. However, the potential cross talk between these two GTPases is unclear. Here, we show that the Rab7 effector PLEKHM1 simultaneously binds Rab7 and Arl8b, bringing about clustering and fusion of late endosomes and lysosomes. We show that the N-terminal RUN domain of PLEKHM1 is necessary and sufficient for interaction with Arl8b and its subsequent localization to lysosomes. Notably, we also demonstrate that Arl8b mediates recruitment of HOPS complex to PLEKHM1-positive vesicle contact sites. Consequently, Arl8b binding to PLEKHM1 is required for its function in delivery and, therefore, degradation of endocytic and autophagic cargo in lysosomes. Finally, we also show that PLEKHM1 competes with SKIP for Arl8b binding, which dictates lysosome positioning. These findings suggest that Arl8b, along with its effectors, orchestrates lysosomal transport and fusion.
© 2017 Marwaha et al.
Figures
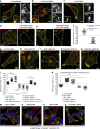
Figure 1.
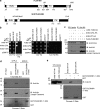
PLEKHM1 directly binds to Arl8b via its N-terminal RUN domain–containing region. (a) Domain architecture of PLEKHM1 and SKIP/PLEKHM2. (b) Yeast two-hybrid assay. Cotransformants were spotted on -Leu-Trp and -Leu-Trp-His media to confirm viability and interactions, respectively. (c) FLAG-PLEKHM1 was cotransfected with different forms of Arl8b-HA into HEK293T cells; lysates were immunoprecipitated (IP) with anti–HA antibody resin, and the precipitates were immunoblotted (IB) with the indicated antibodies. (d and e) GST and GST-PLEKHM1 (1–300) proteins were immobilized on glutathione (GSH) resin and incubated either with His-Arl8b in the presence of GTPγS or GDPβS or with HEK293T cell lysates expressing either Arl8b WT-HA or Arl8b T34N-HA. The precipitates were immunoblotted with anti–His (d) or anti–HA (e) antibodies. Ponceau S stain was done to visualize purified protein. LIR, LC3/GABARAP interaction; PH, pleckstrin homology; WD/WE, tryptophan-acidic.
Figure 2.
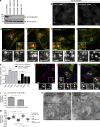
PLEKHM1 colocalizes with Rab7 and Arl8b and promotes perinuclear clustering of lysosomes. (a) Lysates from indicated siRNA treatments and from PLEKHM1 KO-HeLa cells were immunoblotted (IB) with anti-PLEKHM1 antibody for assessing the knockdown efficiency and α-tubulin as the loading control. (b and c) Immunofluorescence depicting the specificity of PLEKHM1 antibody in HeLa cells treated with control- and PLEKHM1-siRNA. (d–h) Representative confocal micrographs of HeLa cells showing endogenous staining of PLEKHM1 with different endocytic markers, and the Pearson’s correlation coefficient (PC) for PLEKHM1 is quantified (n = 3; 25–30 cells analyzed per experiment). (i–k) Representative confocal micrographs of HeLa cells transfected with Arl8b-HA alone or cotransfected with GFP-PLEKHM1 or -NΔ300 PLEKHM1, respectively, and stained for LAMP1. (l) Colocalization of WT and NΔ300 PLEKHM1 with Arl8b was assessed by measuring the PC (n = 3; 75 cells analyzed per experiment). (m) Quantification of perinuclear index of LAMP1+ compartments in HeLa cells transfected with indicated plasmids (n = 3; 15–18 cells analyzed per experiment). (n) Representative immunogold EM image of HeLa cells cotransfected with GFP-PLEKHM1 and Arl8b-HA and labeled with 10- and 15-nm gold particles, respectively. Boxed region is magnified on the right (Bar, 100 nm). Arrowheads mark colocalized pixels. Data represent mean ± SEM (n.s., not significant; **, P < 0.01; ****, P < 0.0001; Student’s t test). Bars: (main) 10 µm; (insets) 2 µm.
Figure 3.
Conserved basic residues within the RUN domain of PLEKHM1 are important for its interaction with Arl8b. (a) Yeast two-hybrid assay. Cotransformants were spotted on -Leu-Trp and -Leu-Trp-His media to confirm viability and interactions, respectively. AD, GAL4 activation domain; BD, GAL4-DNA binding domain. (b) Dot-blot assay: GST alone or GST-PLEKHM1 (1–300) or indicated point mutants were spotted on nitrocellulose membrane and incubated with His-Arl8b or His-Rab7. The interaction was analyzed by immunoblotting (IB) with anti–His antibody. Proteins were visualized by Coomassie staining. (c and d) Representative confocal images showing HeLa cells transfected with GFP-PLEKHM1 or GFP-PLEKHM1 (HRR→A) and immunostained for Arl8. Yellow arrowheads mark colocalized pixels, and white arrowheads mark peripheral Arl8b+-lysosomes. (e) PC quantification of WT or mutant PLEKHM1 with LAMP1 and Rab7 (n = 3; 30 cells analyzed per experiment). (f) Arl8b-HA was cotransfected with FLAG-PLEKHM1 (WT) or HRR→A mutant in HEK293T cells. The lysates were immunoprecipitated (IP) using anti–HA antibody resin and immunoblotted using the indicated antibodies. (g) Immunoblot of a GST pulldown assay using HEK293T cell lysates expressing FLAG-PLEKHM1 (WT) or -Arl8b-binding–defective mutants of PLEKHM1 incubated with GST-Rab7 bound to GSH resin. GST-Rab7 protein was visualized by Ponceau S staining. (h and i) Representative confocal panels showing LAMP1 staining in HeLa cells cotransfected with Arl8b-GFP and FLAG-PLEKHM1 (WT) or HRR→A mutant. LAMP1 staining is shown in insets. (j) Mean size of LAMP1+ compartments in HeLa cells cotransfected with indicated PLEKHM1 plasmid and Arl8b-GFP (n = 3; 25 cells analyzed per experiment). Data represent mean ± SEM (n.s., not significant; *, P < 0.05; ****, P < 0.0001; Student’s t test). Bars: (main) 10 µm; (insets) 2 µm.
Figure 4.
Arl8b is required for PLEKHM1 association with lysosomes and for its ability to promote clustering of LEs and lysosomes. (a) Control- and Arl8b-siRNA (#1 and #2)–treated HeLa cell lysates were immunoblotted (IB) with anti–Arl8 antibody for assessing the knockdown efficiency and α-tubulin as the loading control. The asterisk and arrowhead denote Arl8a and Arl8b protein bands, respectively. (b–e) Representative confocal micrographs depicting the localization of GFP-PLEKHM1 with dextran-647–loaded lysosomes in indicated siRNA treatments and Arl8b siRNA-rescued HeLa cells. Arrowheads mark colocalized pixels. (f–h) Representative confocal micrographs of HeLa cells treated with control- or Arl8b-siRNAs and transfected with GFP-PLEKHM1 followed by immunostaining for Rab7. Arrowheads mark colocalized pixels. (i) PC was calculated as a measure of colocalization of PLEKHM1 with dextran-647–loaded lysosomes or with Rab7 in control siRNA- and Arl8b siRNA-treated HeLa cells (n = 3; 30 cells analyzed per experiment). (j–m) Representative confocal micrographs of HeLa cells expressing GFP-PLEKHM1 and stained for LAMP1 in indicated siRNA treatments and Arl8b siRNA-rescued HeLa cells. Arrowheads mark colocalized pixels. (n) Mean size of PLEKHM1+ compartments in indicated siRNA treatments and Arl8b siRNA-rescued HeLa cells (n = 3; 10–18 cells analyzed per experiment). Data represent mean ± SEM (***, P < 0.001; ****, P < 0.0001; Student’s t test). Bars: (main) 10 µm; (insets) 2 µm.
Figure 5.
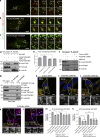
PLEKHM1 acts as a multivalent adaptor that promotes physical interaction between Rab7 and Arl8b. (a–c) Live-cell imaging was performed on cells expressing GFP-Rab7 and Arl8b-tomato along with either FLAG-PLEKHM1 or FLAG-NΔ300 PLEKHM1. The yellow arrowhead depicts kiss-and-run events in a and clustered enlarged endolysosomes in b, respectively. Rab7- and Arl8b-positive punctate structures that do not fuse in c are marked by white and yellow arrowheads. (d) HEK293T cell lysates expressing HA-Rab7 alone or coexpressed with either FLAG-PLEKHM1 (WT) or FLAG-PLEKHM1 (HRR→A) were immunoprecipitated (IP) with anti–HA antibody resin and immunoblotted (IB) using the indicated antibodies. (e) PC of Arl8 and Rab7 immunostained in HeLa cells transfected with indicated plasmids (n = 3; 30 cells analyzed per experiment). (f) Arl8b-HA was transfected in control- or PLEKHM1-siRNA–treated HEK293T cells. The lysates were immunoprecipitated with anti–HA antibody resin and immunoblotted using the indicated antibodies. (g) Lysates of WT- and PLEKHM1 KO-HeLa cells were immunoprecipitated with anti–Rab7 antibody resin and immunoblotted with the indicated antibodies. (h–j) Representative confocal images of HeLa cells treated with control siRNA or PLEKHM1 siRNAs and immunostained with anti–Arl8 and anti–Rab7 antibodies. Arrowheads mark colocalized pixels, and the nucleus was stained using DAPI. (k and l) Representative confocal micrographs of PLEKHM1 siRNA–treated HeLa cells expressing siRNA-resistant GFP-PLEKHM1 (WT) or GFP-PLEKHM1 (HRR→A) and immunostained for Arl8 and Rab7. In the insets, yellow arrowheads mark colocalized pixels. (m and n) PC and MC were calculated for Arl8 and Rab7 colocalization in indicated siRNA treatments of HeLa cells (n = 3; 30 cells analyzed per experiment). Data represent mean ± SEM (n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; Student’s t test). Bars: (main) 10 µm; (insets) 2 µm.
Figure 6.
Arl8b recruits the HOPS complex to Rab7-PLEKHM1–positive endosomes. (a) Lysates from HEK293T cells treated with control- or Arl8b-siRNA and expressing FLAG-PLEKHM1 were IP with anti-FLAG Abs-resin and IB with indicated antibodies. (b–i) Representative confocal micrographs of HeLa cells treated with either control- or Arl8b-siRNA and expressing FLAG-PLEKHM1 (WT) alone or coexpressed with siRNA resistant Arl8b-tomato and stained for Vps41 or Vps18. (j and k) Colocalization of FLAG-PLEKHM1 with Vps41 or Vps18 was quantified by measuring PC in indicated siRNA-treated HeLa cells (n = 3; 30 cells analyzed per experiment). (l) Western blot of GST-pulldown assay using GST-PLEKHM1 (1–300) as bait incubated with lysates from either WT- or Arl8b KO-HeLa cells with increasing concentration of His-Arl8b protein and immunoblotted (IB) with the indicated antibodies. (m) GST-pulldown assay using semipurified TAP–HOPS complex isolated from HeLa cells incubated with either GST or GST-PLEKHM1 (1–300), His-Arl8b, and excess GTP or GDPβS. (n) Lysates of HEK293T cells treated with either control- or Arl8b-siRNA followed by cotransfection with FLAG-PLEKHM1 and HA-Rab7 were subjected to immunoprecipitation (IP) with anti–HA antibody resin and immunoblotted with the indicated antibodies. Data represent mean ± SEM (****, P < 0.0001; Student’s t test). Bars: (main) 10 µm; (insets) 2 µm.
Figure 7.
Binding to Arl8b is necessary for PLEKHM1 function in regulating endocytic cargo trafficking to lysosomes. (a) Schematic illustrating the uptake and further processing of DQ-BSA, an endocytic cargo in the cells. (b–e) Representative confocal images of HeLa cells treated with indicated siRNAs and subjected to DQ-BSA uptake for 6 h. The cells were then fixed and analyzed for DQ-BSA fluorescence. (f) Measurement of fold change in the fluorescence intensity of DQ-BSA from 1h to 6 h (n = 3; 50 cells analyzed per experiment). (g–j) Representative confocal micrographs of HeLa cells treated with the indicated siRNAs and transfected with either siRNA-resistant PLEKHM1 (WT) or siRNA-resistant PLEKHM1 (HRR→A) construct and subjected to DQ-BSA uptake for 6 h. (k) Quantification of DQ-BSA trafficking in HeLa cells treated with indicated siRNAs and transfected with either siRNA-resistant PLEKHM1 (WT) or siRNA-resistant PLEKHM1 (HRR→A) construct (n = 3; 50 cells analyzed per experiment). (l) Western blot of mature cathepsin B and D levels in control- or PLEKHM1-siRNA–treated HeLa cells. (m) PC was measured for cathepsin D, and LAMP1 colocalization in control- or PLEKHM1-siRNA–treated HeLa cells (n = 3; 30 cells per experiment). (n) HeLa cells treated with control- or PLEKHM1-siRNA were incubated for 1 h in growth medium supplemented with cathepsin L substrate, and fluorescence intensity was measured by flow cytometry (n = 3; 10,000 cells analyzed per experiment). Data represent mean ± SEM (n.s., not significant; *, P < 0.05; **, P < 0.01; Student’s t test). Bars, 10 µm.
Figure 8.
PLEKHM1 binds Arl8b to mediate autophagosome–lysosome fusion. (a) U2OS cells were transfected with vector alone (control), FLAG-PLEKHM1 (WT), -PLEKHM1 (HRR→A), or -NΔ300 PLEKHM1 constructs and subjected to 2 h of starvation using EBSS media in the presence or absence of Baf A1. Lysates from these cell types were immunoblotted (IB) with the indicated antibodies. (b–d) Representative confocal micrographs of HeLa cells expressing ptf-LC3B alone, cotransfected with FLAG-PLEKHM1 (WT) or FLAG-PLEKHM1 (HRR→A), and starved for 2 h in EBSS. Red-only punctate structures in magnified insets represent autolysosomes marked by white arrowheads, and yellow punctate structures represent autophagosomes marked by yellow arrowheads. (e) Yeast two-hybrid assay. Cotransformants were spotted on -Leu-Trp and -Leu-Trp-His media to confirm viability and interactions, respectively. (f) U2OS cells treated with the indicated siRNAs were given Baf A1 treatment in normal growth medium. Lysates were immunoblotted with the indicated antibodies. The levels of LC3B-II normalized to α-tubulin were quantified using densitometric analysis as shown. (g) U2OS cells treated with indicated siRNAs were further subjected to the following treatments: normal growth medium or starvation in EBSS media for 2 h with or without Baf A1. The lysates were immunoblotted for the indicated antibodies. Bars: (main) 10 µm; (insets) 2 µm.
Figure 9.
PLEKHM1 and SKIP play opposing roles in regulating lysosome positioning. (a–c) Representative confocal images of HeLa cells transfected with vector, FLAG-PLEKHM1, or FLAG-SKIP and immunostained for Arl8 and LAMP1. (d and e) Representative confocal images of HeLa cells immunostained for Arl8 and PLEKHM1 or SKIP. Only cut-mask image of the colocalized pixels eliminating background and individual pixels are shown on the right. (f) Perinuclear index of colocalized Arl8/PLEKHM1 or Arl8/SKIP pixels were calculated (n = 3; 15–20 cells analyzed per experiment). (g–k) Representative confocal images of HeLa cells treated with indicated siRNAs and stained for Arl8 and LAMP1. (l) PI of LAMP1+-compartments in HeLa cells transfected with indicated siRNAs and siRNA-resistant constructs (n = 3; 14–19 cells analyzed per experiment). (m–p) Representative confocal micrographs of HeLa cells treated with the indicated siRNAs followed by 2-h incubation in acetate Ringer’s solution, pH 6.9, and immunostained for LAMP1 to mark lysosomes. To mark the cell boundary, actin staining was performed using phalloidin and the nucleus was stained using DAPI. (q) Quantification of perinuclear index in HeLa cells treated with indicated siRNAs followed by 2-h incubation in acetate Ringer’s solution (n = 3; 10–18 cells analyzed per experiment). Data represent mean ± SEM (****, P < 0.0001; Student’s t test). Bars: (main) 10 µm; (insets) 2 µm.
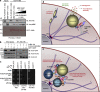
Figure 10.
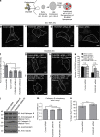
PLEKHM1 and SKIP compete for binding to Arl8b via their respective RUN domains. (a) Immunoblot (IB) of competition assay done using GST-Arl8b as bait and incubated with His-PLEKHM1 (1–300) and increasing concentrations of MBP-SKIP (1–300). (b) Immunoblot of an immunoprecipitation (IP) assay using HEK293T cells lysates coexpressing Arl8b-HA and FLAG-PLEKHM1 with increasing amounts of GFP-SKIP. (c) Yeast three-hybrid assay. Cotransformants were spotted on -Leu-Trp-Met medium to check for viability and on -Leu-Trp-His+2X Met and -Leu-Trp-His-Met media to test the interaction and competition, respectively. (d and e) Proposed model of lysosome distribution and function regulation by small GTPase Arl8b and its effectors, PLEKHM1 and SKIP. SKIP interacts with Arl8b via its RUN domain, further recruiting kinesin motor that drives anterograde lysosome motility, which is implicated in regulating cellular processes like cell migration/invasion and focal adhesion assembly. Here, we report PLEKHM1 as a dual effector of Rab7 and Arl8b that simultaneously binds these GTPases, bringing about clustering and fusion of LEs and lysosomes. PLEKHM1 also binds to LC3 and promotes autolysosome formation.
Similar articles
- SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport.
Jongsma ML, Bakker J, Cabukusta B, Liv N, van Elsland D, Fermie J, Akkermans JL, Kuijl C, van der Zanden SY, Janssen L, Hoogzaad D, van der Kant R, Wijdeven RH, Klumperman J, Berlin I, Neefjes J. Jongsma ML, et al. EMBO J. 2020 Mar 16;39(6):e102301. doi: 10.15252/embj.2019102301. Epub 2020 Feb 21. EMBO J. 2020. PMID: 32080880 Free PMC article. - PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I. McEwan DG, et al. Mol Cell. 2015 Jan 8;57(1):39-54. doi: 10.1016/j.molcel.2014.11.006. Epub 2014 Dec 11. Mol Cell. 2015. PMID: 25498145 - RUFY1 binds Arl8b and mediates endosome-to-TGN CI-M6PR retrieval for cargo sorting to lysosomes.
Rawat S, Chatterjee D, Marwaha R, Charak G, Kumar G, Shaw S, Khatter D, Sharma S, de Heus C, Liv N, Klumperman J, Tuli A, Sharma M. Rawat S, et al. J Cell Biol. 2023 Jan 2;222(1):e202108001. doi: 10.1083/jcb.202108001. Epub 2022 Oct 25. J Cell Biol. 2023. PMID: 36282215 Free PMC article. - Rab7: role of its protein interaction cascades in endo-lysosomal traffic.
Wang T, Ming Z, Xiaochun W, Hong W. Wang T, et al. Cell Signal. 2011 Mar;23(3):516-21. doi: 10.1016/j.cellsig.2010.09.012. Epub 2010 Sep 21. Cell Signal. 2011. PMID: 20851765 Review. - CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion.
Balderhaar HJ, Ungermann C. Balderhaar HJ, et al. J Cell Sci. 2013 Mar 15;126(Pt 6):1307-16. doi: 10.1242/jcs.107805. J Cell Sci. 2013. PMID: 23645161 Review.
Cited by
- HTT (huntingtin) and RAB7 co-migrate retrogradely on a signaling LAMP1-containing late endosome during axonal injury.
Krzystek TJ, White JA, Rathnayake R, Thurston L, Hoffmar-Glennon H, Li Y, Gunawardena S. Krzystek TJ, et al. Autophagy. 2023 Apr;19(4):1199-1220. doi: 10.1080/15548627.2022.2119351. Epub 2022 Sep 9. Autophagy. 2023. PMID: 36048753 Free PMC article. - Polyphosphate is an extracellular signal that can facilitate bacterial survival in eukaryotic cells.
Rijal R, Cadena LA, Smith MR, Carr JF, Gomer RH. Rijal R, et al. Proc Natl Acad Sci U S A. 2020 Dec 15;117(50):31923-31934. doi: 10.1073/pnas.2012009117. Epub 2020 Dec 2. Proc Natl Acad Sci U S A. 2020. PMID: 33268492 Free PMC article. - Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology.
Settembre C, Perera RM. Settembre C, et al. Nat Rev Mol Cell Biol. 2024 Mar;25(3):223-245. doi: 10.1038/s41580-023-00676-x. Epub 2023 Nov 24. Nat Rev Mol Cell Biol. 2024. PMID: 38001393 Review. - A BORC-dependent molecular pathway for vesiculation of cell corpse phagolysosomes.
Fazeli G, Levin-Konigsberg R, Bassik MC, Stigloher C, Wehman AM. Fazeli G, et al. Curr Biol. 2023 Feb 27;33(4):607-621.e7. doi: 10.1016/j.cub.2022.12.041. Epub 2023 Jan 17. Curr Biol. 2023. PMID: 36652947 Free PMC article. - One Disease, Many Genes: Implications for the Treatment of Osteopetroses.
Penna S, Capo V, Palagano E, Sobacchi C, Villa A. Penna S, et al. Front Endocrinol (Lausanne). 2019 Feb 19;10:85. doi: 10.3389/fendo.2019.00085. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 30837952 Free PMC article. Review.
References
- Dykes S.S., Gray A.L., Coleman D.T., Saxena M., Stephens C.A., Carroll J.L., Pruitt K., and Cardelli J.A.. 2016. The Arf-like GTPase Arl8b is essential for three-dimensional invasive growth of prostate cancer in vitro and xenograft formation and growth in vivo. Oncotarget. 7:31037–31052. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases