SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription - PubMed (original) (raw)
SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription
Maximilian F Blank et al. Nucleic Acids Res. 2017.
Abstract
SIRT7 is an NAD+-dependent protein deacetylase that regulates cell growth and proliferation. Previous studies have shown that SIRT7 is required for RNA polymerase I (Pol I) transcription and pre-rRNA processing. Here, we took a proteomic approach to identify novel molecular targets and characterize the role of SIRT7 in non-nucleolar processes. We show that SIRT7 interacts with numerous proteins involved in transcriptional regulation and RNA metabolism, the majority of interactions requiring ongoing transcription. In addition to its role in Pol I transcription, we found that SIRT7 also regulates transcription of snoRNAs and mRNAs. Mechanistically, SIRT7 promotes the release of P-TEFb from the inactive 7SK snRNP complex and deacetylates CDK9, a subunit of the elongation factor P-TEFb, which activates transcription by phosphorylating serine 2 within the C-terminal domain (CTD) of Pol II. SIRT7 counteracts GCN5-directed acetylation of lysine 48 within the catalytic domain of CDK9, deacetylation promoting CTD phosphorylation and transcription elongation.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Figure 1.
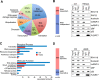
RNA-dependent and -independent SIRT7–protein interactions. (A) Identification of SIRT7-associated proteins. Flag/HA-tagged SIRT7 was purified from HEK293T cells by sequential affinity immunoprecipitation and interacting proteins were analyzed by mass spectrometry. The pie diagram shows functional classification of SIRT7-associated proteins (n = 357). RPs: ribosomal proteins. See also Supplementary Figures S1A, S1B and Supplementary Table S2. (B) RNase A treatment alters the SIRT7 interactome. Flag/HA-tagged SIRT7 was sequentially immunoprecipitated using lysates from HEK293T cells treated with RNase A or left untreated. Interacting proteins were analyzed by mass spectrometry. Left: Percentage of RNase A-sensitive and -insensitive SIRT7–protein interactions. A decrease in the ratio of at least 25% of peptides after RNase treatment was considered as RNA-dependent interaction. Right: Validation of selected SIRT7–protein interactions by co-immunoprecipitation and western blotting. See also Supplementary Figures S1C-E and Supplementary Table S2. (C) AMD treatment alters the SIRT7 interactome. Flag/HA-tagged SIRT7 was purified from untreated or AMD-treated HEK293T cells and interacting proteins were analyzed by mass spectrometry. A decrease in the ratio of at least 25% of peptides after AMD treatment was considered as AMD-sensitive interaction. Gene ontology analysis of proteins associated with Flag/HA-SIRT7 in HEK293T cells treated with AMD (300 ng/ml, 3 h) or left untreated (n = 181). See also Supplementary Figure S1F and Supplementary Table S2. (D) Changes of the SIRT7 interactome upon AMD treatment. Left: Diagram showing AMD-sensitive and insensitive interactions detected by mass spectrometry. Right: Validation of selected AMD-sensitive and -insensitive interactions by co-immunoprecipitation and western blotting.
Figure 2.
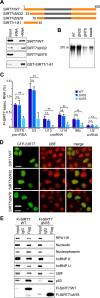
The N-terminal region of SIRT7 mediates interactions with RNA and proteins. (A) The N-terminal part of SIRT7 is required for RNA binding. Nuclear lysates from HEK293T cells expressing Flag-tagged SIRT7 (WT), SIRT7/ΔN32 or SIRT7/ΔN78 were incubated with streptavidin-coated Dynabeads (-RNA) or with Dynabeads containing 5΄ETS-RNA (+RNA). Bound SIRT7 was analyzed on immunoblots (upper panels). Alternatively, bead-bound RNA was incubated with GST-tagged SIRT7/1-81, and binding was monitored with anti-GST antibodies (bottom panel). A scheme illustrating the domain structure of SIRT7 and the deletion mutants is shown above. See also Supplementary Figures S2A and S2B. (B) Pull-down assay showing impaired binding of SIRT7/ΔN78 to RNA. Bead-bound Flag-SIRT7 or the indicated deletion mutants were incubated with radiolabeled 5΄ETS-RNA (+10/+389) and SIRT7-associated RNA was analyzed by gel electrophoresis and PhosphorImaging. RNA bound to beads-only served as a negative control (beads). See also Supplementary Figure S2C. (C) RNA-immunoprecipitation (CLIP) showing that the N-terminal region of SIRT7 mediates RNA binding in vivo. UV-crosslinked Flag-SIRT7-RNA complexes were captured on anti-Flag beads, and co-precipitated RNA was analyzed by RT-qPCR. The percentage of precipitated RNA relative to input RNA is shown. Error bars denote means ±SD (n = 3) (*P < 0.05, **P < 0.01, n.s.: not significant). (D) The N-terminal part is required for nucleolar localization of SIRT7. Direct fluorescence showing the cellular localization of GFP-tagged SIRT7, SIRT7/ΔN32 and SIRT7/ΔN78. Indirect immunofluorescence and direct GFP fluorescence analysis was done as described (13). Nucleoli were stained with anti-UBF antibodies. Scale bar, 10 μm. (E) The N-terminal region of SIRT7 mediates protein interactions. Flag-SIRT7 or mutant ΔN78 were immunoprecipitated and co-precipitated proteins were visualized on immunoblots.
Figure 3.
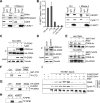
Deacetylation of CDK9 promotes CTD-phosphorylation of Pol II. (A) SIRT7 interacts with Pol II and CDK9. Co-immunoprecipitation experiment showing the association of Flag-SIRT7 with RPB1 and CDK9 in the absence or presence of RNase A. Pol II was monitored with antibodies specific to hypo- and hyperphosphorylated RPB1 (antibody N-20), or with antibodies against phospho-Ser2-CTD. See also Supplementary Figures S3A and S3B. (B) SIRT7 interacts with the 7SK snRNP complex. Left panel: CLIP-qPCR showing the association of Flag-SIRT7 with pre-rRNA (5΄ETS+300/+400 and ITS2) and 7SK RNA, U2 snRNA and actin mRNA serving as negative controls. The bars represent mean values ±SD from three independent experiments (*P < 0.05, **P < 0.01). Right panel: Co-immunoprecipitation experiment showing the association of Flag-SIRT7 with HEXIM1 and cyclin T1 in the absence or presence of RNase A. (C) CDK9 is acetylated by GCN5. HA-CDK9 was immunopurified from HEK293T cells expressing HA-CDK9 and from cells cells co-expressing HA-CDK9 and Flag-GCN5. Where indicated, cells were treated for 5 h with 10 mM nicotinamide (NAM). The level of Flag-GCN5 and acetylation of HA-CDK9 was monitored on western blots. (D) SIRT7 counteracts GCN5-mediated acetylation of CDK9 in vivo. Western blot showing acetylation of HA-CDK9 in SIRT7-depleted HEK293T cells, cells transfected with non-targeting siRNA serving as control. To augment CDK9 acetylation, Flag-GCN5 was co-expressed with HA-CDK9. The expression level of Flag-GCN5 and depletion of SIRT7 by siRNA was monitored on immunoblots. (E) SIRT7 deacetylates CDK9 in vitro. HA-CDK9 was immunopurified from HEK293T cells overexpressing Flag-GCN5 and incubated with Flag-SIRT7/WT (upper panel) or Flag-SIRT7/ΔN78 (lower panels) in the presence or absence of NAD+ (2 mM). Acetylation was monitored on western blots using anti-acetyl-lysine antibodies. See also Supplementary Figures S3C and S3D. (F) Acetylation inhibits CDK9 activity. In vitro kinase assay using GST-CTD as substrate for HA-CDK9 isolated from HEK293T cells expressing HA-CDK9 alone or co-expressing Flag-GCN5 with or without NAM treatment (10 mM, 5 h). Labeling of GST-CTD with γ[32P]ATP was monitored by SDS-PAGE and PhosphorImaging. See also Supplementary Figures S3E and S3F. (G) Acetylation of lysine 48 impairs the CTD kinase activity of CDK9. The enzymatic activity of HA-CDK9 (WT) and point mutants K48R and K48Q was assayed in vitro using immunopurified Flag-RPB1 as substrate. Phosphorylation was monitored by SDS-PAGE and PhosphorImaging. See also Supplementary Figure S3G. (H) Knockdown of SIRT7 impairs CTD phosphorylation at Ser2. In-vitro kinase assay showing Flag-RPB1 phosphorylation by HA-CDK9 purified from SIRT7-depleted HEK293T cells or from cells transfected with non-targeting siRNAs. See also Supplementary Figure S3H. (I) SIRT7 promotes release of P-TEFb from the inactive P-TEFb/7SK snRNP complex. P-TEFb/7SK snRNP complexes were precipitated with anti-HEXIM1 antibodies and bead-bound complexes were incubated with two amounts (1- and 2-fold, see Supplementary Figure S3I) of Flag-SIRT7/WT or Flag-SIRT7/H187Y in the presence or absence of 2 mM NAD+. Release of P-TEFb was monitored on western blots using antibodies against CDK9 and cyclin T1. The bottom panel shows equal amounts of immobilized 7SK/HEXIM1 complex. See also Supplementary Figure S3I.
Figure 4.
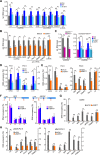
SIRT7 activates transcription of Pol II genes. (A) RT-qPCR analysis of snoRNAs, pre-mRNAs and U1 snRNA from untransfected HEK293T cells and cells overexpressing Flag-SIRT7 (WT) or SIRT7/H187Y. The level of individual RNAs was normalized to actin mRNA. Bars represent means ± SD (n = 3) (*P < 0.05, **P < 0.01). See also Supplementary Figure S4B. (B) snoRNA and pre-mRNA levels are decreased in SIRT7-deficient cells. HEK293T cells were transfected with non-targeting siRNA or SIRT7-specific siRNA. RNA levels were measured by RT-qPCR and normalized to actin mRNA. Bars represent means ± SD (n = 3) (*P < 0.05, **P < 0.01). See also Supplementary Figure S4B. (C) Ectopic SIRT7 rescues downregulation of U3 and U13 snoRNA in SIRT7-knockout cells. HEK293T/SIRT7−/− cells were transfected with Flag-tagged SIRT7/WT, SIRT7/H187Y or SIRT7/ΔN78 and snoRNA levels were monitored by RT-qPCR. The bars represent means ±SD (n = 3) (*P < 0.05, **P < 0.01). See also Supplementary Figure S4C. (D) Overepression of SIRT7 increases the association of the transcription machinery with target genes. ChIP-qPCR monitoring Pol I and Pol II occupancy at selected target genes after overexpression of Flag-SIRT7 (WT) or mutant SIRT7/H187Y. Antibodies against RPA116 (Pol I) and RPB1 (Pol II, N20) were used for ChIP. Bars represent means ± SD (n = 3) (*P < 0.05, **P < 0.01). See also Supplementary Figure S4A. (E) ChIP-qPCR monitoring occupancy of Pol I (anti-RPA116) and Pol II (anti-RPB1, N20) at selected target genes after shRNA- (left panel) or siRNA-(right panels) mediated depletion of SIRT7. Cells transfected with non-targeting shRNA/siRNA served as control. The bars represent means ± SD (n = 3) (*P < 0.05, **P < 0.01). See also Supplementary Figure S4A and S4D. (F) SIRT7 and CDK9 occupancy is enriched at the promoter of target genes. ChIPs monitoring occupancy of endogenous CDK9 or stably expressed Flag-HA-SIRT7 at different regions of Brf2 and JMJD4 using antibodies against CDK9 or the Flag epitope. The bars represent mean values ± SD (n = 3). The scheme depicts the position of the amplified regions. See also Supplementary Figure S4E. (G) Knockdown of SIRT7 impairs CDK9 occupancy at target genes. ChIP-qPCR monitoring occupancy of endogenous CDK9 at target genes upon siRNA-mediated knockdown of SIRT7, cells transfected with non-targeting siRNA serving as control. Bars represent means ± SD (n = 3) (*P < 0.05, **P < 0.01). See also Supplementary Figures S4A and S4F. (H) Knockdown of SIRT7 abolishes Pol II occupancy at selected target genes. ChIPs showing occupancy of Pol II phosphorylated at CTD-Ser5 and CTD-Ser2 in cells transfected with SIRT7-specific siRNA or with non-targeting siRNA using antibodies against pSer5-CTD and pSer2-CTD. Bars represent means ± SD (n = 3) (*P <0.05, **P < 0.01). See also Supplementary Figures S4A and S4F.
Figure 5.
Model illustrating the role of SIRT7 in Pol II transcription. SIRT7-mediated deacetylation promotes the release of CDK9/cyclin T1 from the inactive P-TEFb/7SK snRNP complex and activates P-TEFb by deacetylation of CDK9, which leads to increased CTD-Ser2 phosphorylation and transcriptional processivity.
Similar articles
- Release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein (snRNP) activates hexamethylene bisacetamide-inducible protein (HEXIM1) transcription.
Liu P, Xiang Y, Fujinaga K, Bartholomeeusen K, Nilson KA, Price DH, Peterlin BM. Liu P, et al. J Biol Chem. 2014 Apr 4;289(14):9918-25. doi: 10.1074/jbc.M113.539015. Epub 2014 Feb 10. J Biol Chem. 2014. PMID: 24515107 Free PMC article. - Regulation of P-TEFb elongation complex activity by CDK9 acetylation.
Fu J, Yoon HG, Qin J, Wong J. Fu J, et al. Mol Cell Biol. 2007 Jul;27(13):4641-51. doi: 10.1128/MCB.00857-06. Epub 2007 Apr 23. Mol Cell Biol. 2007. PMID: 17452463 Free PMC article. - Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.
C Quaresma AJ, Bugai A, Barboric M. C Quaresma AJ, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review. - Cellular control of gene expression by T-type cyclin/CDK9 complexes.
Garriga J, Graña X. Garriga J, et al. Gene. 2004 Aug 4;337:15-23. doi: 10.1016/j.gene.2004.05.007. Gene. 2004. PMID: 15276198 Review.
Cited by
- The Mammalian and Yeast A49 and A34 Heterodimers: Homologous but Not the Same.
McNamar R, Rothblum K, Rothblum LI. McNamar R, et al. Genes (Basel). 2021 Apr 22;12(5):620. doi: 10.3390/genes12050620. Genes (Basel). 2021. PMID: 33921963 Free PMC article. Review. - The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy.
Li G, Tian Y, Zhu WG. Li G, et al. Front Cell Dev Biol. 2020 Sep 29;8:576946. doi: 10.3389/fcell.2020.576946. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33117804 Free PMC article. Review. - Molecular Sentinels: Unveiling the Role of Sirtuins in Prostate Cancer Progression.
Chouhan S, Muhammad N, Usmani D, Khan TH, Kumar A. Chouhan S, et al. Int J Mol Sci. 2024 Dec 28;26(1):183. doi: 10.3390/ijms26010183. Int J Mol Sci. 2024. PMID: 39796040 Free PMC article. Review. - DDX3X interacts with SIRT7 to promote PD-L1 expression to facilitate PDAC progression.
Zhao T, Zhu H, Zou T, Zhao S, Zhou L, Ni M, Liu F, Zhu H, Dou X, Di J, Xu B, Wang L, Zou X. Zhao T, et al. Oncogenesis. 2024 Feb 5;13(1):8. doi: 10.1038/s41389-024-00509-2. Oncogenesis. 2024. PMID: 38316768 Free PMC article. - The Emerging Role of SIRT7 in Glucose and Lipid Metabolism.
Yamagata K, Mizumoto T, Yoshizawa T. Yamagata K, et al. Cells. 2023 Dec 25;13(1):48. doi: 10.3390/cells13010048. Cells. 2023. PMID: 38201252 Free PMC article. Review.
References
- Kiran S., Anwar T., Kiran M., Ramakrishna G.. Sirtuin 7 in cell proliferation, stress and disease: rise of the seventh Sirtuin!. Cell. Signal. 2015; 27:673–682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous