A Ca2+-stimulated exosome release pathway in cancer cells is regulated by Munc13-4 - PubMed (original) (raw)
A Ca2+-stimulated exosome release pathway in cancer cells is regulated by Munc13-4
Scott W Messenger et al. J Cell Biol. 2018.
Erratum in
- Correction: A Ca2+-stimulated exosome release pathway in cancer cells is regulated by Munc13-4.
Messenger SW, Woo SS, Sun Z, Martin TFJ. Messenger SW, et al. J Cell Biol. 2019 Apr 1;218(4):1423. doi: 10.1083/jcb.20171013203042019c. Epub 2019 Mar 9. J Cell Biol. 2019. PMID: 30852488 Free PMC article. No abstract available.
Abstract
Cancer cells secrete copious amounts of exosomes, and elevated intracellular Ca2+ is critical for tumor progression and metastasis, but the underlying cellular mechanisms are unknown. Munc13-4 is a Ca2+-dependent SNAP receptor- and Rab-binding protein required for Ca2+-dependent membrane fusion. Here we show that acute elevation of Ca2+ in cancer cells stimulated a fivefold increase in CD63+, CD9+, and ALIX+ exosome release that was eliminated by Munc13-4 knockdown and not restored by Ca2+ binding-deficient Munc13-4 mutants. Direct imaging of CD63-pHluorin exosome release confirmed its Munc13-4 dependence. Depletion of Munc13-4 in highly aggressive breast carcinoma MDA-MB-231 cells reduced the size of CD63+ multivesicular bodies (MVBs), indicating a role for Munc13-4 in MVB maturation. Munc13-4 used a Rab11-dependent trafficking pathway to generate MVBs competent for exosome release. Membrane type 1 matrix metalloproteinase trafficking to MVBs by a Rab11-dependent pathway was also Munc13-4 dependent, and Munc13-4 depletion reduced extracellular matrix degradation. These studies identify a novel Ca2+- and Munc13-4-dependent pathway that underlies increased exosome release by cancer cells.
© 2018 Messenger et al.
Figures
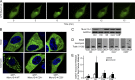
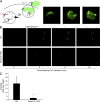
Figure 1.
Munc13-4 KD strongly impairs exosome release. (A) SDS-PAGE Western blot of indicated proteins in MDA-MB-231 cells after stable expression of shRNA for Munc13-4 or Rab27a or a scrambled control (Ctrl). (B) Culture medium from MDA-MB-231 cells either untreated or stimulated with 1.25 µM ionomycin for 30 min was centrifuged at 1,000 g to remove cellular debris and 10,000 g to remove large extracellular vesicles. (C) The resulting 10,000-g supernatant was filtered onto a nitrocellulose membrane and analyzed for CD63, CD9, ALIX, and GM130 content by antibody blotting. (D) Quantification of CD63, CD9, and ALIX blots in C are shown as exosome release as a percentage of total cellular material with mean values ± standard error (SE) for n ≥ 3. *, P < 0.05 for comparison with corresponding control samples. (E) Panc-1 or A549 cells were left untreated or were treated with TGFβ-1 for 24 h. Indicated proteins were detected by SDS-PAGE Western blot. (F) Panc-1 cells were left untreated or were treated with TGFβ-1 for 24 h, and Munc13-4 levels were determined by immunofluorescence. TGFβ-1–treated cells exhibited a mesenchymal morphology. Bars, 5 μm. (G) A549 cells stably expressing control shRNA (Ctrl) or Munc13-4 shRNA were left untreated (Untr) or were treated with TGFβ-1 for 24 h, and SDS-PAGE Western blotting for indicated proteins was conducted. (H) Culture media supernatants (as in B) from A549 cells that were either untreated or were stimulated with 1.25 µM ionomycin for 30 min were filtered onto nitrocellulose membrane and analyzed for CD63 and GM130. (I) Quantification of CD63+ exosome release shown as a percentage of total cellular material with mean values ± SE for n = 5. *, P < 0.05; **, P < 0.01 for comparison with corresponding control samples.
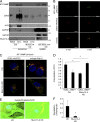
Figure 2.
Munc13-4 translocation to membrane is Ca2+ dependent. (A) Live-cell epifluorescence imaging of GFP-Munc13-4 in MDA-MB-231 cells at indicated times after ionomycin stimulation. See Video 1. (B) MDA-MB-231 cells expressing wild-type GFP-Munc13-4, GFP-Munc13-4 C2A*, or GFP-Munc13-4 C2B* either left untreated or stimulated with 1.25 µM ionomycin for 5 min were fixed and imaged by confocal microscopy. (C) Indicated proteins were detected by SDS-PAGE and Western blotting of lysates with Munc13-4 antibody from MDA-MB-231 cells stably expressing control shRNA (Ctrl) or shRNA targeting Munc13-4 (KD), or Munc13-4 KD cells rescued with shRNA-resistant wild-type Munc13-4, Munc13-4 C2A*, or Munc13-4 C2B*. (D) Culture media supernatants (as in Fig. 1 B) from MDA-MB-231 cells as in C either untreated or stimulated with 1.25 µM ionomycin for 30 min were filtered onto membrane and analyzed for CD63 or GM130. (E) Quantification of CD63+ exosome release (from Fig 1 D) shown as mean values ± SE for n = 3. *, P < 0.05 for comparison between ionomycin-treated and basal. Bars, 5 μm.
Figure 3.
Munc13-4 regulates MVB size. (A and B) MDA-MB-231 cells stably expressing control shRNA (Ctrl) or Munc13-4 shRNA were fixed for immunofluorescence detection of endogenous CD63 (red) and LAMP1 (green) by confocal microscopy (A) or SIM (B). Bars, 5 μm. (C) Mean diameter of CD63+ structures in each indicated cell type imaged by confocal or SIM shown as mean values ± SE for three cells/group from three separate preparations (*, P < 0.05). (D) SIM showing orthogonal (top) and 3D reconstruction view of CD63 (red) and LAMP1 (green) in MDA-MB-231 cells stably expressing control shRNA (Ctrl) or Munc13-4. Bars, 5 μm. (E) 3D reconstruction of SIM images from Ctrl MDA-MB-231 cell in D of a single MVB labeling CD63 (red) and LAMP1 (green). Linescan of indicated channels is shown on the bottom. Bars, 1 µm.
Figure 4.
Munc13-4 recruitment to recycling endosomes is dependent on Rab11a. (A) MDA-MB-231 cells immunolabeled for endogenous CD63 (red) and Munc13-4 (green) by confocal microscopy. (B) MDA-MB-231 cells expressing GFP-Munc13-4 and mCherry-Rab27a, mCherry-Rab27b, or mApple-Rab11a imaged by confocal microscopy. (C) Percentage of cells with Pearson’s correlation coefficient for GFP versus mCherry/mApple >0.7. (D) MDA-MB-231 cells stably expressing control shRNA (Ctrl) or Rab11a shRNA and GFP-Munc13-4 were left untreated or were stimulated with 1.25 µM ionomycin for 5 min, fixed, and imaged by confocal microscopy. (E) SDS-PAGE Western blot of indicated proteins in cells stably expressing control shRNA (Ctrl) or Rab11a shRNA. (F) Culture media supernatants (as in Fig. 1 B) from control (Ctrl) or MDA-MB-231 cells stimulated with 1.25 µM ionomycin for 30 min were filtered onto membrane and immunoblotted for CD63 and GM130. (G) Quantification of CD63+ exosome release as percentage of cellular total indicated as mean values ± SE for n = 5. **, P < 0.01 for comparison of ionomycin-treated samples. Bars, 5 μm.
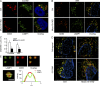
Figure 5.
Munc13-4 function in exosome release is dependent on Rab11a. (A) MDA-MB-231 cells expressing GFP-Rab11, GFP-Rab11 Q70L, or GFP-Rab11 S25N were immunolabeled for myc-Munc13-4 (red) and endogenous CD63 (cyan). Bar, 5 µm. (B) Pearson’s correlation coefficient for GFP-Rab11 versus CD63 immunolocalization. Mean values ± SE (15 cells/group from three separate preparations) are shown. *, P < 0.05; **, P < 0.01. (C) MDA-MB-231 cells expressing GFP, GFP-Rab11, or GFP-Rab11 S25N were immunoblotted for the indicated proteins. Arrow indicates GFP-Rab11; lower band in upper panel corresponds to Rab11. (D) Cell medium supernatants (as in Fig. 1 B) from untreated MDA-MB-231 cells or cells treated with 1.25 µM ionomycin were filtered onto membrane and analyzed for CD63 and GM130. (E) Quantification of CD63 exosome release as percentage of total cellular shown as mean values ± SE (n = 3) with *, P < 0.05 for comparison to GFP-alone samples. (F) Model for the generation of secretion-competent MVBs as arising from the transient fusion of Rab11+ endosomes with MVB precursors, which requires Munc13-4, Ca2+, and Rab11a. MT1-MMP (green bar) is found in the recycling endosome (RE) and delivered to the MVB dependent on Munc13-4 for incorporation to ILVs and exosome release. The secretion-competent MVBs generated acquire components for Ca2+-dependent fusion with the plasma membrane. Bars, 5 μm.
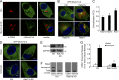
Figure 6.
Direct TIRF imaging of exosome secretion dependent on Munc13-4. (A) pH-sensitive pHluorin (pHl) was inserted into loop 1 of CD63, which localizes to both the limiting membrane and ILVs of MVBs. Initially quenched at the low pH in the MVB, CD63-pHluorin increases fluorescence upon fusion of MVBs with the plasma membrane. (B) MDA-MB-231 cells stably expressing control shRNA (Ctrl, upper row) or Munc13-4 shRNA (Munc13-4 KD, lower row) were transfected with CD63-pHluorin plasmid, stimulated with 1.25 µM ionomycin (at zero time), and imaged for 15 min by TIRF microscopy. The surface of single cells is shown. In the upper row, ionomycin treatment elicited the release of brightened CD63-pHluorin–containing ILVs released into TIRF field. Bar, 5 µm. Inset shows one of the MVBs releasing ILVs as exosomes. Bar, 1 µm. See Videos 2 and 3. (C) Intensity of CD63-pHluorin signal is quantified as fold increase over initial fluorescent value reported as mean values ± SE (≥9 cells/group; *, P < 0.05).
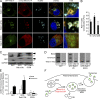
Figure 7.
ECM degradation by exosomal MT1-MMP is dependent on Munc13-4. (A) Exosomes (100,000-g pellet) and 400 µg cell lysate were prepared from MDA-MB-231 cells stably expressing control shRNA (Ctrl) or Munc13-4 shRNA (Munc13-4 KD) and analyzed for indicated proteins by SDS-PAGE Western blotting. Exosomes secreted basally or from cells stimulated with 1.25 µM ionomycin for 1 h were compared. (B) MDA-MB-231 cells expressing CD63-mKATE2 and MT1-MMP-pHluorin were imaged by TIRF microscopy before (0 min) and after treatment with 1.25 µM ionomycin for 5 min. (C) Control or Munc13-4 KD cells expressing MT1-MMP-pHluorin and CD63-mKATE2 or mApple-Rab11a were fixed, permeabilized to brighten pHluorin, and imaged by confocal microscopy. (D) Pearson’s correlation coefficient for pHluorin versus mKATE2 or mApple is indicated as mean values ± SE (15 cells/group from three separate preparations are shown). (E) MDA-MB-231 cells stably expressing control shRNA (Ctrl) or Munc13-4 shRNA were grown on cover slides coated with Oregon-Green-Gelatin for 16 h and labeled with phalloidin and DAPI. Zoomed images from representative cells are shown. (F) Gelatin degradation was calculated as a percentage of clearing of gelatin from the larger field per cell, shown as mean values ± SE (nine fields of view [5–25 cells]/group from three separate preparations). *, P < 0.05. Bars, 5 μm.
Similar articles
- Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer.
Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Han QF, et al. Mol Cancer. 2022 Nov 1;21(1):207. doi: 10.1186/s12943-022-01671-0. Mol Cancer. 2022. PMID: 36320056 Free PMC article. Review. - PIKfyve inhibition increases exosome release and induces secretory autophagy.
Hessvik NP, Øverbye A, Brech A, Torgersen ML, Jakobsen IS, Sandvig K, Llorente A. Hessvik NP, et al. Cell Mol Life Sci. 2016 Dec;73(24):4717-4737. doi: 10.1007/s00018-016-2309-8. Epub 2016 Jul 20. Cell Mol Life Sci. 2016. PMID: 27438886 Free PMC article. - Live-Imaging Detection of Multivesicular Body-Plasma Membrane Fusion and Exosome Release in Cultured Primary Neurons.
Pescosolido MF, Ouyang Q, Liu JS, Morrow EM. Pescosolido MF, et al. Methods Mol Biol. 2023;2683:213-220. doi: 10.1007/978-1-0716-3287-1_17. Methods Mol Biol. 2023. PMID: 37300778 - CD63 Regulates Epstein-Barr Virus LMP1 Exosomal Packaging, Enhancement of Vesicle Production, and Noncanonical NF-κB Signaling.
Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG Jr. Hurwitz SN, et al. J Virol. 2017 Feb 14;91(5):e02251-16. doi: 10.1128/JVI.02251-16. Print 2017 Mar 1. J Virol. 2017. PMID: 27974566 Free PMC article. - Current knowledge on exosome biogenesis and release.
Hessvik NP, Llorente A. Hessvik NP, et al. Cell Mol Life Sci. 2018 Jan;75(2):193-208. doi: 10.1007/s00018-017-2595-9. Epub 2017 Jul 21. Cell Mol Life Sci. 2018. PMID: 28733901 Free PMC article. Review.
Cited by
- Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer.
Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Han QF, et al. Mol Cancer. 2022 Nov 1;21(1):207. doi: 10.1186/s12943-022-01671-0. Mol Cancer. 2022. PMID: 36320056 Free PMC article. Review. - Extracellular Vesicles as Delivery Vehicles for Therapeutic Nucleic Acids in Cancer Gene Therapy: Progress and Challenges.
Du R, Wang C, Zhu L, Yang Y. Du R, et al. Pharmaceutics. 2022 Oct 19;14(10):2236. doi: 10.3390/pharmaceutics14102236. Pharmaceutics. 2022. PMID: 36297672 Free PMC article. Review. - Identification of immune-infiltrated hub genes as potential biomarkers of Moyamoya disease by bioinformatics analysis.
Jin F, Duan C. Jin F, et al. Orphanet J Rare Dis. 2022 Feb 23;17(1):80. doi: 10.1186/s13023-022-02238-4. Orphanet J Rare Dis. 2022. PMID: 35197088 Free PMC article. - Exosomes, a New Star for Targeted Delivery.
Chen H, Wang L, Zeng X, Schwarz H, Nanda HS, Peng X, Zhou Y. Chen H, et al. Front Cell Dev Biol. 2021 Oct 8;9:751079. doi: 10.3389/fcell.2021.751079. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34692704 Free PMC article. Review. - Tumor-derived exosomes (TDEs): How to avoid the sting in the tail.
Wan M, Ning B, Spiegel S, Lyon CJ, Hu TY. Wan M, et al. Med Res Rev. 2020 Jan;40(1):385-412. doi: 10.1002/med.21623. Epub 2019 Jul 18. Med Res Rev. 2020. PMID: 31318078 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- F32 CA200024/CA/NCI NIH HHS/United States
- R01 DK025861/DK/NIDDK NIH HHS/United States
- R37 DK025861/DK/NIDDK NIH HHS/United States
- T32 DK007665/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous