TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis - PubMed (original) (raw)
TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis
Wei Wang et al. Autophagy. 2018.
Abstract
TRIM37 gene mutations cause mulibrey (muscle-liver-brain-eye) nanism, a severe growth disorder with prenatal onset. Although TRIM37 depletion normally induces apoptosis, patients with TRIM37 mutations have a high risk of developing tumors, suggesting that there may be an alternative pro-survival mechanism for TRIM37-deficient tumor cells. We find that TRIM37 interacts with MTOR and RRAGB proteins, enhances the MTOR-RRAGB interaction and promotes lysosomal localization of MTOR, thereby activating amino acid-stimulated MTORC1 signaling. In response to loss of TRIM37 functions, phosphorylation of TFEB is significantly reduced, resulting in its translocation into the nucleus enabling its transcriptional activation of genes involved in lysosome biogenesis and macroautophagy/autophagy. The enhanced autophagy depends on the inhibition of MTORC1 signaling and may serve as an alternative mechanism to survive the loss of TRIM37 functions. Our study unveils a positive role of TRIM37 in regulating the MTORC1-TFEB axis and provides mechanistic insights into the pathogenesis of mulibrey nanism, as well as potential therapeutic treatment.
Abbreviations: ACTB: actin beta; ATG: autophagy related; CASP3: caspase3; CLEAR: coordinated lysosomal expression and regulation; CQ: chloroquine; CTS: cathepsin proteases; CTSL: cathepsin L; EIF4EBP1: eukaryotic translation initiation factor 4E binding protein 1; LAMP1: lysosomal associated membrane protein 1; LAMP2: lysosomal associated membrane protein 2; LMNB1: lamin B1; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MTOR: mechanistic target of rapamycin kinase; MTORC1: MTOR complex 1; mulibrey: muscle-liver-brain-eye; NAC: N-acetyl-L-cysteine; PARP1: poly(ADP-ribose) polymerase 1; RAP2A: member of RAS oncogene family; RHEB: Ras homolog enriched in brain; ROS: reactive oxygen species; RPS6KB1: ribosomal protein S6 kinase B1; RRAGB: Ras related GTP binding B; SQSTM1: sequestosome 1; TFEB: transcription factor EB; TRIM37: tripartite motif containing 37.
Keywords: Autophagy; MTORC1; TFEB; TRIM37; lysosome.
Figures
Figure 1.
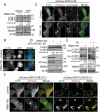
TRIM37 depletion induces autophagy. (a) HepG2, HEK 293T control, and TRIM37 KD cells grown in normal growth media were collected for detection of the proteins via western blotting using the indicated antibodies. Actin beta (ACTB) was used as a loading control. (b) HepG2 control and TRIM37 KD cells were immuno-stained with anti-LC3B antibody to detect the localization of endogenous LC3B proteins using regular immuno-fluorescence microscopy. (c) TRIM37 was depleted (TRIM37 KD) in HepG2 cells stably expressing mCherry-EGFP-LC3B. Cells grown in normal growth media were incubated with LysoTracker Blue (0.2 μM) for 30 min before fixation with 4% paraformaldehyde and subsequent confocal image acquisition. (d) HEK 293T control and TRIM37 KD cells were transfected with two siRNAs for ATG7 and ATG16L1. Cells were collected after 72 h for protein detection with the indicated antibodies. (e) HepG2 control and TRIM37 KD cells were treated with CQ (10 μM) for 6 h prior to western blot detection of proteins shown. s.e. short exposure; l.e. long exposure. (f) HepG2 control and TRIM37 KD cells stably expressing mCherry-EGFP-LC3B were treated with (+) or without (-) CQ (10 μM) for 6 h before confocal image acquisition. Scale bars: 10 μm. The intensities of LC3B-II were quantified and presented as the ratio of LC3B-II:ACTB below the LC3B blots as ± SEM (n = 3) from 3 independent experiments. The LC3B-II:ACTB value in control cells was set as 1.
Figure 2.
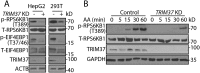
TRIM37 depletion inhibits amino acid-stimulated MTORC1 signaling. (a) HepG2, HEK 293T control, and TRIM37 KD cells were cultured in normal growth media and collected for protein detection via western blotting with the indicated antibodies. T-RPS6KB1 and T-EIF4EBP1 indicate total RPS6KB1 and EIF4EBP1, respectively. ACTB was used as a loading control. (b) HepG2 control and TRIM37 KD cells were incubated in media depleted of amino acids (AA) [38] for 2 h and stimulated with amino acid-containing media for the indicated times, before proteins were detected by western blots. GAPDH was used as a loading control.
Figure 3.
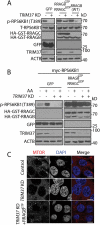
TRIM37 interacts with MTOR and RRAGB proteins and its depletion inhibits MTOR-RRAGB interaction and lysosomal distribution of MTOR. (a) HEK 293T cells were transfected with FLAG-MTOR and TRIM37-GFP constructs. After 24 h, equal amounts of cell lysates were collected for immunoprecipitation (IP) with control IgG (Ctrl) or FLAG antibody, followed by protein detection with GFP and MTOR antibodies. (b and c) HEK 293T cells were transfected with HA-GST-RRAGB and TRIM37-GFP constructs. After 24 h, cells lysates were collected (normal). Alternatively, cells were starved for amino acid for 2 h (AA-) and stimulated with amino acid-containing media for 1 h (AA+). Equal amounts of cell lysates were collected for immuno-precipitation (IP) with control IgG (Ctrl) or HA antibody. GFP and HA-HRP antibodies were used to detect TRIM37-GFP and HA-GST-RRAGB, respectively. (d and e) HepG2 control and TRIM37 KD cells were incubated in amino acid-depleted media [38] for 2 h (AA-) and then stimulated with amino acid-containing media (AA+) for 15 min. Nuclei were stained with DAPI. (d) Cells were immuno-stained with MTOR antibody. (e) Cells stimulated with AA (15 min) were co-stained with MTOR and LAMP2 antibodies. The images were acquired by confocal microscopy. Scale bars: 10 μm. (F) HEK 293T control and TRIM37 KD cells were transfected with FLAG-MTOR and HA-GST-RRAGB constructs. After 24 h, equal amounts of cell lysates were collected for immuno-precipitation (IP) with control IgG (Ctrl) or HA antibody, followed by protein detection with the indicated antibodies. s.e. short exposure; l.e. long exposure.
Figure 4.
TRIM37 promotes MTOR signaling through RRAGB. (a) HepG2 control and TRIM37 KD cells were transfected with constructs encoding GFP only, RRAGBGTP (HA-GST-RRAGBQ99L) plus RRAGCGDP (HA-GST-RRAGCS75L), or HA-GST-RRAGB (WT). Cells were collected after 24 h of transfection, before proteins were detected by western blots. (b) HepG2 control and TRIM37 KD cells were transfected with constructs encoding myc-RPS6KB1, together with GFP or RRAGBGTP plus RRAGCGDP . After 24 h, cells were incubated with amino acid-depletion media for 2 h, followed by stimulation with amino acid-containing media for 15 min, before proteins were detected by western blots. (c) HepG2 control, TRIM37 KD cells and TRIM37 KD cells expressing FLAG-RRAGBGTP were immunostained with MTOR antibody. Nuclei were stained with DAPI. The images were acquired by confocal microscopy. Scale bars: 10 μm.
Figure 5.
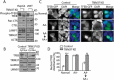
TRIM37 depletion reduces TFEB phosphorylation and induces TFEB nuclear translocation. (a) HepG2, HEK 293T control, and TRIM37 KD cells grown in normal growth media were collected for protein detection with the indicated antibodies. The smeared bands represent TFEB phosphorylation (phospho-TFEB). Pan-cathepsin (pan-CTS) antibody recognizes cathepsin family proteins. ACTB is used as a loading control. (b) HepG2 control and TRIM37 KD cells were fractionated into cytoplasm (Cyt) and nucleus (Nuc), followed by protein detection with the indicated antibodies. GAPDH and LMNB1 (lamin B1) were used as control proteins for cytoplasmic and nuclear fractions, respectively. (c and d) The TFEB-GFP expression construct was transfected into HepG2 control or TRIM37 KD cells. After 24 h, cells were incubated in amino acid-free media for 2 h (AA-), followed by incubation in amino acid-containing media (AA+) for 2 h. Normal, shows cells grown in normal growth media. Nuclei were stained with DAPI. Scale bars: 5 μm. (d) Quantification of cells shown in (c). Approximately 20–30 cells expressing TFEB-GFP were obtained and the percentages of cells with predominantly nuclear TFEB-GFP distributions were quantified. The results are presented as mean ± SEM (n = 3) based on 3 independent experiments. **, p < 0.01; ***, p < 0.001 (Student’s t test).
Figure 6.
TRIM37 depletion induces nuclear translocation of MiTF/TFE family proteins and activates the CLEAR network. (a and b) HepG2 control and TRIM37 KD cells were transfected with constructs expressing TFEB-GFP, TFE3-GFP, MITF-A-GFP, or MITF-D-GFP. Images were taken after 24 h. Scale bars: 5 μm. (b) Quantification of cells in (a). Approximately 20–30 GFP-positive cells were obtained and the percentages of cells with predominantly nuclear distributions of these GFP fusion proteins were quantified. The results are presented as mean ± SEM (n = 3) based on 3 independent experiments. ***, P < 0.001 (Student’s t test). (c) Real-time PCR analysis of the autophagy genes and lysosome biogenesis genes in HepG2 control and TRIM37 KD cells. Data are shown as mean ± SEM, n = 3. *: p < 0.05, **: p < 0.01, ***: p < 0.001 (Student’s _t_-test).
Figure 7.
RRAGBGTP expression rescues MTORC1 inhibition and compromises autophagy induction by TRIM37 depletion. (a–c) TRIM37 was silenced in HepG2 cells stably expressing FLAG-RAP2A or FLAG-RRAGBGTP (FLAG-RRAGBQ99L). (a) These cells in normal growth media were collected for protein detection with the indicated antibodies. ACTB is used as a loading control. (b) The cells were also transfected with the TFEB-GFP construct. After 24 h, the localization was analyzed by microscopy. Nuclei were stained with DAPI. Scale bars: 5 μm. (c) Quantification of cells shown in (b). Approximately 20–30 cells expressing TFEB-GFP were obtained and the percentages of cells with predominantly nuclear TFEB-GFP distributions were quantified. The results are presented as mean ± SEM (n = 3) based on 3 independent experiments. ***, p < 0.001 (Student’s t test). FLAG-RAP2A was used as a negative control protein, as described previously [24].
Figure 8.
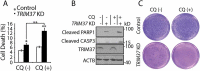
Survival of TRIM37 KD cells may depend on lysosomal functions. (a and b) HepG2 control and TRIM37 KD cells were treated without (-) or with (+) CQ (10 μM) for 16 h. (a) Cells were collected for PI staining to analyze the percentages of cell death. Data from triplicate samples in each group are presented as mean ± SEM and representative of 3 independent experiments; *: p < 0. 05; **: p < 0. 01; (Student’s t-test). (b) Cells were also collected for protein detection with the indicated antibodies. (c) HepG2 control and TRIM37 KD cells were grown in soft agar in the absence (-) or presence (+) of CQ (10 μM). The pictures were taken after 2 weeks.
Similar articles
- Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis.
Wang S, Ni HM, Chao X, Wang H, Bridges B, Kumer S, Schmitt T, Mareninova O, Gukovskaya A, De Lisle RC, Ballabio A, Pacher P, Ding WX. Wang S, et al. Autophagy. 2019 Nov;15(11):1954-1969. doi: 10.1080/15548627.2019.1596486. Epub 2019 Mar 30. Autophagy. 2019. PMID: 30894069 Free PMC article. - TFEB-driven endocytosis coordinates MTORC1 signaling and autophagy.
Nnah IC, Wang B, Saqcena C, Weber GF, Bonder EM, Bagley D, De Cegli R, Napolitano G, Medina DL, Ballabio A, Dobrowolski R. Nnah IC, et al. Autophagy. 2019 Jan;15(1):151-164. doi: 10.1080/15548627.2018.1511504. Epub 2018 Sep 10. Autophagy. 2019. PMID: 30145926 Free PMC article. - Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration.
Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V, Vezzoli G, Tedesco B, Meroni M, Messi E, Piccolella M, Galbiati M, Garrè M, Morelli E, Vaccari T, Poletti A. Rusmini P, et al. Autophagy. 2019 Apr;15(4):631-651. doi: 10.1080/15548627.2018.1535292. Epub 2018 Nov 5. Autophagy. 2019. PMID: 30335591 Free PMC article. - How autophagy controls the intestinal epithelial barrier.
Foerster EG, Mukherjee T, Cabral-Fernandes L, Rocha JDB, Girardin SE, Philpott DJ. Foerster EG, et al. Autophagy. 2022 Jan;18(1):86-103. doi: 10.1080/15548627.2021.1909406. Epub 2021 Apr 27. Autophagy. 2022. PMID: 33906557 Free PMC article. Review. - TFEB: a double-edged sword for tumor metastasis.
Hu JH, Li SY, Yu LH, Guan ZR, Jiang YP, Hu D, Wang HJ, Zhao LP, Zhou ZH, Yan YX, Xie T, Huang ZH, Lou JS. Hu JH, et al. J Mol Med (Berl). 2023 Aug;101(8):917-929. doi: 10.1007/s00109-023-02337-0. Epub 2023 Jun 17. J Mol Med (Berl). 2023. PMID: 37328669 Review.
Cited by
- The Tripartite Nexus: Autophagy, Cancer, and Tripartite Motif-Containing Protein Family Members.
Mandell MA, Saha B, Thompson TA. Mandell MA, et al. Front Pharmacol. 2020 Mar 11;11:308. doi: 10.3389/fphar.2020.00308. eCollection 2020. Front Pharmacol. 2020. PMID: 32226386 Free PMC article. Review. - TRIM37 exacerbates hepatic ischemia/reperfusion injury by facilitating IKKγ translocation.
Yang H, Huang Z, Luo Y, Lei D, Yan P, Shen A, Liu W, Li D, Wu Z. Yang H, et al. Mol Med. 2023 May 8;29(1):62. doi: 10.1186/s10020-023-00653-2. Mol Med. 2023. PMID: 37158850 Free PMC article. - TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism.
Brigant B, Metzinger-Le Meuth V, Rochette J, Metzinger L. Brigant B, et al. Int J Mol Sci. 2018 Dec 24;20(1):67. doi: 10.3390/ijms20010067. Int J Mol Sci. 2018. PMID: 30586926 Free PMC article. Review. - UBE3A deficiency-induced autophagy is associated with activation of AMPK-ULK1 and p53 pathways.
Hao X, Sun J, Zhong L, Baudry M, Bi X. Hao X, et al. Exp Neurol. 2023 May;363:114358. doi: 10.1016/j.expneurol.2023.114358. Epub 2023 Feb 26. Exp Neurol. 2023. PMID: 36849003 Free PMC article. - The ubiquitin ligase TRIM32 promotes the autophagic response to Mycobacterium tuberculosis infection in macrophages.
Romagnoli A, Di Rienzo M, Petruccioli E, Fusco C, Palucci I, Micale L, Mazza T, Delogu G, Merla G, Goletti D, Piacentini M, Fimia GM. Romagnoli A, et al. Cell Death Dis. 2023 Aug 5;14(8):505. doi: 10.1038/s41419-023-06026-1. Cell Death Dis. 2023. PMID: 37543647 Free PMC article.
References
- Avela K, Lipsanen-Nyman M, Idanheimo N, et al. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in mulibrey nanism. Nat Genet. 2000. July;25(3):298–301. PubMed PMID: 10888877. - PubMed
- Karlberg N, Jalanko H, Kallijarvi J, et al. Insulin resistance syndrome in subjects with mutated RING finger protein TRIM37. Diabetes. 2005. December;54(12):3577–3581. PubMed PMID: 16306379. - PubMed
- Karlberg N, Karlberg S, Karikoski R, et al. High frequency of tumours in mulibrey nanism. J Pathol. 2009. June;218(2):163–171. PubMed PMID: 19334051. - PubMed
- Hamalainen RH, Mowat D, Gabbett MT, et al. Wilms’ tumor and novel TRIM37 mutations in an Australian patient with mulibrey nanism. Clin Genet. 2006. December;70(6):473–479. PubMed PMID: 17100991. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous