Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications - PubMed (original) (raw)
Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications
Keith A Jolley et al. Wellcome Open Res. 2018.
Abstract
The PubMLST.org website hosts a collection of open-access, curated databases that integrate population sequence data with provenance and phenotype information for over 100 different microbial species and genera. Although the PubMLST website was conceived as part of the development of the first multi-locus sequence typing (MLST) scheme in 1998 the software it uses, the Bacterial Isolate Genome Sequence database (BIGSdb, published in 2010), enables PubMLST to include all levels of sequence data, from single gene sequences up to and including complete, finished genomes. Here we describe developments in the BIGSdb software made from publication to June 2018 and show how the platform realises microbial population genomics for a wide range of applications. The system is based on the gene-by-gene analysis of microbial genomes, with each deposited sequence annotated and curated to identify the genes present and systematically catalogue their variation. Originally intended as a means of characterising isolates with typing schemes, the synthesis of sequences and records of genetic variation with provenance and phenotype data permits highly scalable (whole genome sequence data for tens of thousands of isolates) means of addressing a wide range of functional questions, including: the prediction of antimicrobial resistance; likely cross-reactivity with vaccine antigens; and the functional activities of different variants that lead to key phenotypes. There are no limitations to the number of sequences, genetic loci, allelic variants or schemes (combinations of loci) that can be included, enabling each database to represent an expanding catalogue of the genetic variation of the population in question. In addition to providing web-accessible analyses and links to third-party analysis and visualisation tools, the BIGSdb software includes a RESTful application programming interface (API) that enables access to all the underlying data for third-party applications and data analysis pipelines.
Keywords: Database; epidemiology; evolution; population annotation; public health.
Conflict of interest statement
No competing interests were disclosed.
Figures
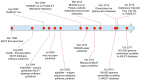
Figure 1.. MLST comes of age: 21 years of population genomics.
The PubMLST website (
) has been running for 15 years, having been established under the pubmlst.org domain in 2003. Its immediate progenitor was the original MLST database set up to support the_Neisseria_ scheme,, the first MLST scheme developed in 1998. The initial role of the site was to host the nomenclature and isolate collection records for typing schemes, but it was rapidly opened to the wider community, hosting schemes of other organisms . Shortly afterwards, other sites began hosting MLST schemes, the most prominent of which was mlst.net , at Imperial College, along with others at the University of Cork, Ireland later migrated to University of Warwick and subsumed within the Enterobase platform , and the Pasteur Institute, Paris, France. Early generations of software developed to support the databases– were limited to specific loci defined for a single typing scheme specified in their configuration. With extensive WGS data in prospect, in 2008 work started on a platform designed to flexibly handle genomic data utilizing any number of loci and typing schemes. The resulting Bacterial Isolate Genome Sequence Database (BIGSdb) platform has been used since then to host databases on PubMLST as well as being used for the databases hosted at the Pasteur Institute. It has been under constant development since. In 2016, the databases hosted on mlst.net were migrated to PubMLST, with the result that most MLST schemes are now hosted using the same platform (the major exceptions being_Salmonella_ and_Escherichia coli_, hosted on Enterobase, although these schemes are mirrored on PubMLST).
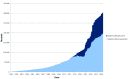
Figure 2.. Submission of isolate records and genomes to the PubMLST species/genus-specific databases over time.
Prior to 2012, most submissions consisted of provenance metadata along with MLST results and a few antigen sequence designations. Since then, the proportion of submissions that include whole genome assemblies has continually increased. The apparent dip in isolates without genomes which occurred around 2014 was due to genome assemblies being added to existing records that had been submitted previously with just MLST results.
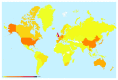
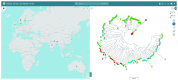
Figure 3.. Global submissions to the PubMLST databases.
Isolate records submitted to individual databases represent almost every country in the world. There are approximately 125 curators handling submissions from over 2000 active data submitters across all the species- and genus-specific databases hosted on PubMLST.
Figure 4.. Analysis pipeline for short read data.
PubMLST and BIGSdb, its underlying genomics platform, links provenance metadata with allelic sequence variation found in corresponding whole genome assemblies or sequences derived from Sanger sequencing reactions. Population annotation, the process of assigning precise variant information for loci across the genomes of large numbers of bacterial isolates, creates a structured dataset that can be used to address a range of biological questions beyond epidemiology.
Figure 5.. Data flow overview.
Individuals may have different and overlapping roles: Users query and analyse data; submitters upload data for nomenclature assignment and inclusion in databases; and curators assign allele and profile identifiers, check metadata, upload genomes and perform allele calling (largely automated with manual oversight). Interaction with the PubMLST BIGSdb databases is via the web interface or RESTful API. Analysis of datasets returned by a query can be performed using integrated tools or forwarded to third party sites using their APIs to upload results in their required formats.
Figure 6.. Extracting typing information from a local genome file.
Typing information can be readily extracted from whole genome sequence assemblies using the sequence query page. (A) Genome assembly contigs are either pasted in to the sequence query form and the required scheme or locus (in this case, MLST) selected. (B) Any locus exact matches are displayed and, if this corresponds to a defined combination of alleles, the profile definition (ST/clonal complex for MLST) is displayed.
Figure 7.. GrapeTree minimum-spanning tree of Neisseria meningitidis genomes (n=12,179) differentiated by cgMLST (1605 loci).
A minimum-spanning tree based on allelic profiles can be generated using isolate records returned from any query and any selected scheme or group of loci. The dataset was selected by searching for species ‘_Neisseria meningitidis_’ with an attached sequence bin size of >2 Mbp, indicative of a complete genome. Nodes are coloured by clonal complex as defined by classical MLST (7 locus), indicating strong concordance between typing schemes. Branches shorter than 150 loci are collapsed.
Figure 8.. Spatio-phylogenetic analysis of the global Dichelobacter nodosus population.
The geographical distribution of clades of_Dichelobacter nodosus_ was demonstrated by analysing all genomes in the database (n=171) using the BIGSdb Microreact plugin with cgMLST loci. This created a concatenated alignment of core genes which was used to generate a Neighbor-joining tree that was automatically uploaded to the Microreact website with accompanying metadata for visualization.
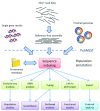
Figure 9.. Relationships of community, tasks, methods and tools supported by PubMLST.
PubMLST links structured bacterial isolate datasets with whole genome sequence data and molecular typing nomenclature to provide a rich resource that can be exploited for a wide range of tasks including surveillance, vaccine development, evolutionary analysis and functional studies.
Similar articles
- BIGSdb: Scalable analysis of bacterial genome variation at the population level.
Jolley KA, Maiden MC. Jolley KA, et al. BMC Bioinformatics. 2010 Dec 10;11:595. doi: 10.1186/1471-2105-11-595. BMC Bioinformatics. 2010. PMID: 21143983 Free PMC article. - A Gene-By-Gene Approach to Bacterial Population Genomics: Whole Genome MLST of Campylobacter.
Sheppard SK, Jolley KA, Maiden MC. Sheppard SK, et al. Genes (Basel). 2012 Apr 12;3(2):261-77. doi: 10.3390/genes3020261. Genes (Basel). 2012. PMID: 24704917 Free PMC article. - MLSTar: automatic multilocus sequence typing of bacterial genomes in R.
Ferrés I, Iraola G. Ferrés I, et al. PeerJ. 2018 Jun 15;6:e5098. doi: 10.7717/peerj.5098. eCollection 2018. PeerJ. 2018. PMID: 29922519 Free PMC article. - A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes.
Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. Bratcher HB, et al. BMC Genomics. 2014 Dec 18;15(1):1138. doi: 10.1186/1471-2164-15-1138. BMC Genomics. 2014. PMID: 25523208 Free PMC article. - Population and Functional Genomics of Neisseria Revealed with Gene-by-Gene Approaches.
Maiden MC, Harrison OB. Maiden MC, et al. J Clin Microbiol. 2016 Aug;54(8):1949-55. doi: 10.1128/JCM.00301-16. Epub 2016 Apr 20. J Clin Microbiol. 2016. PMID: 27098959 Free PMC article. Review.
Cited by
- Four New Sequence Types and Molecular Characteristics of Multidrug-Resistant Escherichia coli Strains from Foods in Thailand.
Thadtapong N, Chaturongakul S, Tangphatsornruang S, Sonthirod C, Ngamwongsatit N, Aunpad R. Thadtapong N, et al. Antibiotics (Basel). 2024 Oct 2;13(10):935. doi: 10.3390/antibiotics13100935. Antibiotics (Basel). 2024. PMID: 39452202 Free PMC article. - Genomic Characterization of 16S rRNA Methyltransferase-Producing Enterobacterales Reveals the Emergence of Klebsiella pneumoniae ST6260 Harboring rmtF, rmtB, _bla_NDM-5, _bla_OXA-232 and _bla_SFO-1 Genes in a Cancer Hospital in Bulgaria.
Sabtcheva S, Stoikov I, Georgieva S, Donchev D, Hodzhev Y, Dobreva E, Christova I, Ivanov IN. Sabtcheva S, et al. Antibiotics (Basel). 2024 Oct 10;13(10):950. doi: 10.3390/antibiotics13100950. Antibiotics (Basel). 2024. PMID: 39452216 Free PMC article. - MGTdb: a web service and database for studying the global and local genomic epidemiology of bacterial pathogens.
Kaur S, Payne M, Luo L, Octavia S, Tanaka MM, Sintchenko V, Lan R. Kaur S, et al. Database (Oxford). 2022 Nov 11;2022:baac094. doi: 10.1093/database/baac094. Database (Oxford). 2022. PMID: 36367311 Free PMC article. - Synthetic DNA and biosecurity: Nuances of predicting pathogenicity and the impetus for novel computational approaches for screening oligonucleotides.
Elworth RAL, Diaz C, Yang J, de Figueiredo P, Ternus K, Treangen T. Elworth RAL, et al. PLoS Pathog. 2020 Aug 6;16(8):e1008649. doi: 10.1371/journal.ppat.1008649. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32760120 Free PMC article. No abstract available. - Burkholderia cepacia Complex Taxon K: Where to Split?
Depoorter E, De Canck E, Peeters C, Wieme AD, Cnockaert M, Zlosnik JEA, LiPuma JJ, Coenye T, Vandamme P. Depoorter E, et al. Front Microbiol. 2020 Jul 14;11:1594. doi: 10.3389/fmicb.2020.01594. eCollection 2020. Front Microbiol. 2020. PMID: 32760373 Free PMC article.
References
Grants and funding
Development of PubMLST and BIGSdb has been supported by a Wellcome Trust Biomedical Resource Grant (104992). Design and implementation of the RESTful API has been further supported by the European Community grant FP7-278864-2 (PathoNgenTrace, http://www.patho-ngen-trace.eu/).
LinkOut - more resources
Full Text Sources
Other Literature Sources