CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines - PubMed (original) (raw)
. 2018 Dec 21:10:247-264.
doi: 10.1016/j.isci.2018.11.029. Epub 2018 Dec 12.
Augustin Luna 2, Mihoko Yamade 3, Lisa Loman 4, Sudhir Varma 4, Margot Sunshine 5, Francesco Iorio 6, Fabricio G Sousa 7, Fathi Elloumi 5, Mirit I Aladjem 4, Anish Thomas 4, Chris Sander 8, Kurt W Kohn 4, Cyril H Benes 9, Mathew Garnett 6, William C Reinhold 4, Yves Pommier 10
Affiliations
- PMID: 30553813
- PMCID: PMC6302245
- DOI: 10.1016/j.isci.2018.11.029
CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines
Vinodh N Rajapakse et al. iScience. 2018.
Abstract
CellMinerCDB provides a web-based resource (https://discover.nci.nih.gov/cellminercdb/) for integrating multiple forms of pharmacological and genomic analyses, and unifying the richest cancer cell line datasets (the NCI-60, NCI-SCLC, Sanger/MGH GDSC, and Broad CCLE/CTRP). CellMinerCDB enables data queries for genomics and gene regulatory network analyses, and exploration of pharmacogenomic determinants and drug signatures. It leverages overlaps of cell lines and drugs across databases to examine reproducibility and expand pathway analyses. We illustrate the value of CellMinerCDB for elucidating gene expression determinants, such as DNA methylation and copy number variations, and highlight complexities in assessing mutational burden. We demonstrate the value of CellMinerCDB in selecting drugs with reproducible activity, expand on the dominant role of SLFN11 for drug response, and present novel response determinants and genomic signatures for topoisomerase inhibitors and schweinfurthins. We also introduce LIX1L as a gene associated with mesenchymal signature and regulation of cellular migration and invasiveness.
Keywords: Bioinformatics; Biological Database; Cancer Systems Biology; Genomics.
Published by Elsevier Inc.
Figures
Graphical abstract
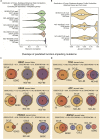
Figure 1
CellMinerCDB Overview (A) CellMinerCDB integrates cancer cell line information from principal resources and provides powerful, user-friendly analysis tools. (B) Summary of molecular and drug activity data for the five data sources currently included in CellMinerCDB. For molecular data types, the numbers indicate the number of genes with a particular data type. GDSC gene-level mutation and methylation data (numbers in red) were prepared from raw data as part of the development of CellMinerCDB. Asterisks indicate molecular data under development, but not publicly available. Protein expression was determined by reverse-phase protein array. (C) Cell line and drug overlaps between data sources. (D) Drug overlaps between data sources. (E) Small cell lung cancer (SCLC) cell line overlaps between data sources. (F) SCLC cell line-tested drug overlaps between data sources.
Figure 2
Molecular Data Reproducibility across Sources Comparison of the available genomic features of the cell lines shared between the CellMinerCDB data sources. Bar plots indicate the median and inter-quartile range. (A) Pearson's correlation distributions for comparable expression (exp), DNA copy number (cop), and DNA methylation (met) data. (B) Jaccard coefficient distributions for comparable binary mutation (mut) data. The Jaccard coefficient for a pair of gene-specific mutation profiles is the ratio of the number of mutated cell lines reported by both sources to the number of mutated lines reported by either source. (C and D) Overlaps of function-impacting mutations as predicted using SIFT/PolyPhen2 for selected tumor suppressor genes and oncogenes. Matched cell line mutation data were binarized by assigning a value of 1 to lines with a homozygous mutation probability greater than a threshold, which was set to 0.3 for (B) and for oncogenes in (C) and to 0.7 for tumor suppressor genes in (D).
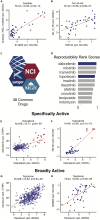
Figure 3
Drug Activity Data Reproducibility (A and B) GDSC versus NCI-60 drug activity data in matched cell lines for (A) oxyphenisatin acetate (acetalax; NSC59687) and (B) MJ-III-65 (LMP744; NSC706744). Each point represents a matched cell line. Red points in (A) indicate triple-negative breast cancer cell lines. (C–H) (C and D) A total of 38 drugs were tested in the NCI-60, GDSC, and CTRP. CCLE was excluded because of its small drug dataset (24 drugs), which is largely included in CTRP. For each of the three inter-source comparisons, drugs were ranked by activity correlation strength (q-value), with ranks scaled between 0 (lowest) and 1 (highest). Specifically active compounds, such as the BRAF inhibitor dabrafenib, show strong correlations based on the response of melanoma lines shown in red (E and F), whereas broadly active compounds, such as the topoisomerase I inhibitor topotecan, show strong correlations based on broad response patterns (G and H). The NCI-60-matched data in (F) and (H) capture the pattern observed with matched data between the larger GDSC and CTRP collections. The full data table excerpted in (D) is shown in Figure S6.
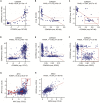
Figure 4
Exploring Gene Expression Determinants Reduced mRNA expression (xai, average log2 intensity) of the cell cycle inhibitor and tumor suppressor CDKN2A (p16) is associated with DNA copy loss (cop) (A) and promoter methylation (met) (B) in the NCI-60 lines. In a subset of NCI-60 lines, enclosed in the red box, (C), DNA copy loss accompanies higher levels of promoter methylation. DNA copy number and promoter methylation data from the CCLE and GDSC, respectively, can be also be visualized over matched cell lines to verify a similar pattern in larger cell line collections (D–F). Note that the corroboration of the NCI-60 regulatory relationships in a far larger and more diverse cell line set is uniquely enabled by CellMinerCDB, which allows gene-level methylation data only available in the GDSC to complement gene-level DNA copy number data only available in the CCLE (for automatically matched cell lines). DNA copy number gain is associated with increased expression (exp, Z score microarray log2 intensity data) of the oncogenes MYC (G) and KRAS (H) in selected CCLE cell lines. In (G), small cell lung cancer lines are indicated in red to highlight a subset potentially derived from MYC-driven tumors (within red box).
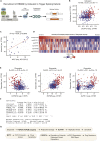
Figure 5
Exploring Drug Response Determinants (A and B) Response to the pre-mRNA splicing inhibitor indisulam versus expression of its target complex component DCAF15 in the CTRP. Drug response in (B) is measured by the activity area above the dose-response curve, with higher values indicating relative drug sensitivity. A report of increased indisulam sensitivity in hematopoietic cell lines (shown in red) with high DCAF15 expression is readily verified (Han et al., 2017). (C) Response to the aurora kinase inhibitor alisertib is associated with increased MYC expression in small cell lung cancer lines (Mollaoglu et al., 2017). (D) Heatmap indicating etoposide drug activity and candidate determinant gene expression in the 100 most sensitive and resistant CTRP cell lines. (E) Scatterplots of etoposide activity versus candidate determinant gene expression in CTRP cell lines, with hematopoietic cell lines shown in red. (F) A statistical summary of a multivariate linear model of etoposide response in the CTRP. (G) A mechanistic scheme indicating how the selected determinants may influence etoposide drug response.
Figure 6
A Multivariate Model of Schweinfurthin A Drug Activity (A) Reproducibility of the data for the two schweinfurthin derivatives tested in the GDSC. (B) Heatmap indicating Schweinfurthin A drug activity and candidate determinant gene expression in the 100 most sensitive and resistant non-hematopoietic GDSC cell lines. (C) A statistical summary of a multivariate linear model of Schweinfurthin A response in the GDSC. (D) Scheme of the proposed molecular pharmacology of the schweinfurthins. Schweinfurthins have been shown to inhibit PI3K/AKT signaling and cell survival by binding oxysterol-binding-protein-related proteins (ORPs) to disrupt _trans_-Golgi network trafficking required for robust pathway activity (Bao et al., 2015). Together with the ORPs OSBP, OSBPL3, and OSBPL10, the other candidate determinants, PLEKHO1 and THEM4, have also been implicated in PI3K/AKT signaling (Liu et al., 2013, Tokuda et al., 2007). Plots and analyses in panels B–D are based on non-hematopoietic GDSC cell lines.
Figure 7
Relating Epithelial Mesenchymal Transition (EMT) Status with Gene Expression to Identify LIX1L as a Novel EMT Gene (A) A 37-gene EMT signature developed in (Kohn et al., 2014) was used to derive a numerical index of EMT status as a weighted sum of cell-line-specific EMT gene expression values (see Transparent Methods for details). Epithelial and mesenchymal statuses are associated with positive and negative index values, respectively. (B) For 821 non-hematopoietic cell lines in the GDSC collection, the EMT index values show a bimodal distribution, which can be modeled as a normal mixture. Cell lines with EMT index values less than (greater than) 1 standard deviation above (below) the putative mesenchymal (epithelial) group mean are annotated as mesenchymal (epithelial). (C) EMT stratification by tissue of origin. (D and E) Expression of LIX1L, a novel mesenchymal gene, is strongly correlated with the EMT index signature. “Epithelial-mesenchymal” lines with intermediate EMT index values are indicated in red. Mesenchymal lines are in blue at the left, and epithelial are in blue at the right. (F) Western blot showing the efficient knockdown of LIX1L in MDA-MB231 cells. (G) Representative image showing increased migration and invasion after LIX1L knockdown. (H) Quantitation of the increased migration and invasion of cells after LIX1L knockdown. Individual experiments are shown as dots. Error bars indicate the standard error of the mean.
Similar articles
- CellMiner Cross-Database (CellMinerCDB) version 1.2: Exploration of patient-derived cancer cell line pharmacogenomics.
Luna A, Elloumi F, Varma S, Wang Y, Rajapakse VN, Aladjem MI, Robert J, Sander C, Pommier Y, Reinhold WC. Luna A, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D1083-D1093. doi: 10.1093/nar/gkaa968. Nucleic Acids Res. 2021. PMID: 33196823 Free PMC article. - CellMinerCDB: NCATS Is a Web-Based Portal Integrating Public Cancer Cell Line Databases for Pharmacogenomic Explorations.
Reinhold WC, Wilson K, Elloumi F, Bradwell KR, Ceribelli M, Varma S, Wang Y, Duveau D, Menon N, Trepel J, Zhang X, Klumpp-Thomas C, Micheal S, Shinn P, Luna A, Thomas C, Pommier Y. Reinhold WC, et al. Cancer Res. 2023 Jun 15;83(12):1941-1952. doi: 10.1158/0008-5472.CAN-22-2996. Cancer Res. 2023. PMID: 37140427 Free PMC article. - RNA Sequencing of the NCI-60: Integration into CellMiner and CellMiner CDB.
Reinhold WC, Varma S, Sunshine M, Elloumi F, Ofori-Atta K, Lee S, Trepel JB, Meltzer PS, Doroshow JH, Pommier Y. Reinhold WC, et al. Cancer Res. 2019 Jul 1;79(13):3514-3524. doi: 10.1158/0008-5472.CAN-18-2047. Epub 2019 May 21. Cancer Res. 2019. PMID: 31113817 Free PMC article. - Using CellMiner 1.6 for Systems Pharmacology and Genomic Analysis of the NCI-60.
Reinhold WC, Sunshine M, Varma S, Doroshow JH, Pommier Y. Reinhold WC, et al. Clin Cancer Res. 2015 Sep 1;21(17):3841-52. doi: 10.1158/1078-0432.CCR-15-0335. Epub 2015 Jun 5. Clin Cancer Res. 2015. PMID: 26048278 Free PMC article. Review. - Evaluating the consistency of large-scale pharmacogenomic studies.
Rahman R, Dhruba SR, Matlock K, De-Niz C, Ghosh S, Pal R. Rahman R, et al. Brief Bioinform. 2019 Sep 27;20(5):1734-1753. doi: 10.1093/bib/bby046. Brief Bioinform. 2019. PMID: 31846027 Free PMC article. Review.
Cited by
- A systematic evaluation of deep learning methods for the prediction of drug synergy in cancer.
Baptista D, Ferreira PG, Rocha M. Baptista D, et al. PLoS Comput Biol. 2023 Mar 23;19(3):e1010200. doi: 10.1371/journal.pcbi.1010200. eCollection 2023 Mar. PLoS Comput Biol. 2023. PMID: 36952569 Free PMC article. - Platinum-Acridine Agents with High Activity in Cancers Expressing the Solute Carrier MATE1 (SLC47A1).
Zhang S, Wu H, Day CS, Bierbach U. Zhang S, et al. ACS Med Chem Lett. 2023 Jul 25;14(8):1122-1128. doi: 10.1021/acsmedchemlett.3c00266. eCollection 2023 Aug 10. ACS Med Chem Lett. 2023. PMID: 37583829 Free PMC article. - An unbiased high-throughput drug screen reveals a potential therapeutic vulnerability in the most lethal molecular subtype of pancreatic cancer.
Pan CH, Otsuka Y, Sridharan B, Woo M, Leiton CV, Babu S, Torrente Gonçalves M, Kawalerski RR, K Bai JD, Chang DK, Biankin AV, Scampavia L, Spicer T, Escobar-Hoyos LF, Shroyer KR. Pan CH, et al. Mol Oncol. 2020 Aug;14(8):1800-1816. doi: 10.1002/1878-0261.12743. Epub 2020 Jul 4. Mol Oncol. 2020. PMID: 32533886 Free PMC article. - CDKN2A-Inactivated Pancreatic Ductal Adenocarcinoma Exhibits Therapeutic Sensitivity to Paclitaxel: A Bioinformatics Study.
Lin JC, Liu TP, Yang PM. Lin JC, et al. J Clin Med. 2020 Dec 12;9(12):4019. doi: 10.3390/jcm9124019. J Clin Med. 2020. PMID: 33322698 Free PMC article. - CellMiner Cross-Database (CellMinerCDB) version 1.2: Exploration of patient-derived cancer cell line pharmacogenomics.
Luna A, Elloumi F, Varma S, Wang Y, Rajapakse VN, Aladjem MI, Robert J, Sander C, Pommier Y, Reinhold WC. Luna A, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D1083-D1093. doi: 10.1093/nar/gkaa968. Nucleic Acids Res. 2021. PMID: 33196823 Free PMC article.
References
- Antony S., Jayaraman M., Laco G., Kohlhagen G., Kohn K.W., Cushman M., Pommier Y. Differential induction of topoisomerase I-DNA cleavage complexes by the indenoisoquinoline MJ-III-65 (NSC 706744) and camptothecin: base sequence analysis and activity against camptothecin-resistant topoisomerases I. Cancer Res. 2003;63:7428–7435. - PubMed
- Bao X., Zheng W., Hata Sugi N., Agarwala K.L., Xu Q., Wang Z., Tendyke K., Lee W., Parent L., Li W. Small molecule schweinfurthins selectively inhibit cancer cell proliferation and mTOR/AKT signaling by interfering with trans-Golgi-network trafficking. Cancer Biol. Ther. 2015;16:589–601. - PMC - PubMed
- Boehm J.S., Golub T.R. An ecosystem of cancer cell line factories to support a cancer dependency map. Nat. Rev. Genet. 2015;16:373–374. - PubMed
Grants and funding
- 102696/WT_/Wellcome Trust/United Kingdom
- F32 CA192901/CA/NCI NIH HHS/United States
- P41 GM103504/GM/NIGMS NIH HHS/United States
- Z01 BC006150/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources