USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7 - PubMed (original) (raw)
. 2019 Mar 29;10(1):1454.
doi: 10.1038/s41467-019-09437-x.
Aysegul Sapmaz 1 2 3, Ilana Berlin 1 2 3, Ruud H Wijdeven 1 2 3, Hans Janssen 1, Rebecca Konietzny 4 5, Jimmy J Akkermans 2 3, Ayse E Erson-Bensan 6, Roman I Koning 3, Benedikt M Kessler 4, Jacques Neefjes 7 8 9, Huib Ovaa 10 11 12
Affiliations
- PMID: 30926795
- PMCID: PMC6440979
- DOI: 10.1038/s41467-019-09437-x
USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7
Aysegul Sapmaz et al. Nat Commun. 2019.
Abstract
The endosomal system is a highly dynamic multifunctional organelle, whose complexity is regulated in part by reversible ubiquitylation. Despite the wide-ranging influence of ubiquitin in endosomal processes, relatively few enzymes utilizing ubiquitin have been described to control endosome integrity and function. Here we reveal the deubiquitylating enzyme (DUB) ubiquitin-specific protease 32 (USP32) as a powerful player in this context. Loss of USP32 inhibits late endosome (LE) transport and recycling of LE cargos, resulting in dispersion and swelling of the late compartment. Using SILAC-based ubiquitome profiling we identify the small GTPase Rab7-the logistical centerpiece of LE biology-as a substrate of USP32. Mechanistic studies reveal that LE transport effector RILP prefers ubiquitylation-deficient Rab7, while retromer-mediated LE recycling benefits from an intact cycle of Rab7 ubiquitylation. Collectively, our observations suggest that reversible ubiquitylation helps switch Rab7 between its various functions, thereby maintaining global spatiotemporal order in the endosomal system.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
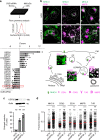
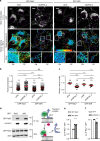
Deubiquitylating enzyme (DUB) screen reveals USP32 as a regulator of endosome biology. a Small interfering RNA (siRNA)-based screen for DUBs affecting major histocompatability class II receptor (MHC-II) surface levels. MelJuSo cells transfected with siRNAs targeting all human DUBs were analyzed for surface expression of peptide-loaded MHC-II by flow cytometry using monoclonal antibodies (L243-Cy3). _Z_-scores of DUBs whose depletion resulted in elevated (Z ≥ 3) or diminished (Z ≤ −3) levels of MHC-II on the cell surface are plotted, n = 3 biologically independent samples. Effect of USP32 depletion is highlighted by a red box. b Effect of USP32 loss on the size and distribution of endosomes. Representative confocal overlays of fixed MelJuSo cells transfected with either control (siCtrl) siRNA or oligo #2 targeting USP32 (siUSP32_2) and immunostained against MHC-II (green) and vesicular markers or cargoes (magenta) are shown. EEA1: early endosome (EE) marker, CD63 late endosome (LE)/multi-vesicular body (MVB) marker, mannose-6-phosphate receptor (M6PR): trans-Golgi network (TGN) cargo. Transferrin receptor (TrfR): recycling endosome (RE) marker; PM: plasma membrane. Cell and nuclear boundaries are demarcated with dashed white lines, scale bars = 10 µm. Zooms Z1–Z3 are placed within a schematic of cargo flow. c Percentage cells harboring enlarged MHC-II-positive vesicles, n = 3 independent experiments. Immunoblot of USP32 protein expression in response to depletion using two independent siRNA oligos (siUSP32_1 and siUSP32_2) is provided with actin as loading control. d Vesicle dispersion expressed as fractional distance of MHC-II pixels (black open circles) along a straight line from the center of nucleus (0) to the PM (1.0). Red lines: mean, n = 2 independent experiments. Total number of cells analyzed per condition appears above each bar/scatter. Bar graphs report mean of independent sample values (black circles), error bars reflect ±s.d. All significant values were calculated using Student’s t test: **p < 0.01, ***p < 0.001, NS = not significant. See also Supplementary Fig. 1
Fig. 2
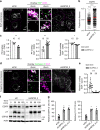
Loss of USP32 disrupts cargo trafficking and lysosomal proteolysis. a–e Effect of USP32 depletion on ligand-mediated trafficking and degradation of epidermal growth factor (EGF) receptor (EGFR). a Representative confocal z-projections of fixed HeLa cells transfected as indicated, starved and stimulated with 100 ng/mL EGF-555 (white) for 120 min. Perinuclear (PN) and peripheral (PP) insets show overlays of EGF (green) with immunostained CD63 (magenta). b EGF-positive pixel distribution expressed as fractional distance along a straight line from center of nucleus (0) to the PM (1.0). Red lines: mean, n = 2 independent experiments. c Colocalization of EGF with CD63 in PN (left), PP (middle), and overall (right) in control cells (siCtrl, white bars) vs. those depleted of USP32 (siUSP32_2, gray bars), n = 2 independent experiments. d Representative confocal images of fixed HeLa cells transfected as indicated, starved and stimulated with EGF-555 (white) for 120 min. PN and PP insets show overlays of EGF (magenta) with immunostained cathepsin D (green). e Colocalization of EGF with cathepsin D in control cells (siCtrl, white bars) vs. those depleted of USP32 (siUSP32_2, gray bars), n = 3 independent experiments. All colocalization plots report Mander’s overlap quantified from multicell images (black circles). Cell and nuclear boundaries are depicted in dashed magenta and white lines, respectively. Scale bars = 10 μm. f, g Effect of USP32 depletion on ligand-induced degradation of EGFR. f Lysates from HeLa cells transfected as indicated, serum starved, and stimulated with EGF (25 ng/mL) for 0, 30, 60, or 120 min were analyzed by immunoblot against total EGFR (rabbit anti-EGFR) and phosphorylated (pY) EGFR (mouse anti-phosphotyrosine 4G10), with actin as a loading control. g Total (left graph, relative to t = 0) and activated (right graph, pY relative to t = 30) EGFR remaining at 120 min following stimulation in control cells (siCtrl) vs. those depleted of USP32 using different siRNA oligos (siUSP32_2 and siUSP32_3 + 4), n = 3 independent experiments. Bar graphs report mean of independent measurements (black circles), error bars reflect ±s.d. Total number of cells analyzed per condition appears above each bar/scatter. All significance calculated using Student’s t test: *p < 0.05, **p < 0.01, and ***p < 0.001. See also Supplementary Fig. 2
Fig. 3
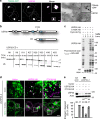
Catalytic activity of USP32 supports endosomal system’s architecture. a Localization of USP32 to the trans-Golgi network (TGN). Left panels: representative confocal images of fixed MelJuSo cells stably expressing TGN46-GFP (green) and immunostained against endogenous USP32 (magenta). Cell and nuclear boundaries are demarcated with dashed white lines, scale bars = 10 µm. Right panels: Electron micrographs of sections co-labeled with anti-USP32 (10 nm gold) and anti-GFP (15 nm gold), scale bar = 0.25 µm. See also Supplementary Fig. 3 and Movies 1–8. b Top panel: schematic representation of USP32 domain organization: EF, calcium-binding domain; DUSP, domain found in ubiquitin-specific proteases (USP); UBL, ubiquitin-like domain; USP, catalytic domain harboring the principal catalytic residue C743. Bottom panel: in vitro cleavage of di-ubiquitin linkages (M1, K6, K11, K27, K29, K33, K48, and K63) by the catalytic domain (CD) of USP32. See also Supplementary Fig. 4. c DUB activity-based probe assay performed on lysates of HEK293T cells expressing USP32-HA or catalytic mutant C743A-HA in the absence (−) or presence (+) of Cy5-Ub-Prg probe. Reaction products were analyzed using in-gel fluorescence scanning followed by immunoblot against HA; * indicates USP32-HA labeling. d, e Rescue of USP32 depletion phenotypes of LE enlargement and dispersion by re-expression of siUSP32_2-resistant USP32-HA vs. C743A-HA relative to empty vector. d Representative confocal images of fixed MelJuSo cells transfected as indicated and immunostained against major histocompatibility class II (MHC-II) (green) and HA (magenta) are shown with the corresponding zooms. Cell and nuclear boundaries are demarcated with dashed white lines. Arrows point to HA-positive puncta juxtaposed to endosomes, scale bars = 10 µm. e Enlargement and dispersion of MHC-II-positive vesicles reported as % cells. Bars depict mean of n = 3 independent experiments (black circles), error bars reflect ±s.d., total number of cells analyzed per condition appears above each bar. Immunoblot against USP32 is shown with actin as a loading control. All significance was calculated using Student’s t test: ***p < 0.001, NS = not significant
Fig. 4
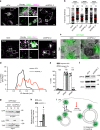
Ubiquitome analysis reveals small GTPase Rab7 as a substrate of USP32. a Schematic representation of stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative mass spectrometry (LC-MS/MS) workflow used to compare ubiquitylated proteomes of b control MelJuSo cells (siCtrl, green) vs. those where USP32 was knocked down (KD, siUSP32_2, magenta) and c HeLa cells overexpressing (OE) USP32-HA (magenta) vs. vector control (green). Cell growth media types: K0R0, light; K4R6, medium; K8R10, heavy. IP: immunoprecipitation, m/z: mass to charge ratio. b, c Volcano plots comparing abundance of detected peptides carrying a GlyGly (GG) modification expressed as Log 2 ratios of b USP32 knockdown (siUSP32_2, KD) vs. control (siCtrl, CTRL), n = 1 SILAC sample set independently validated using label-free quantitation (LFQ) with n = 2 biologically independent samples or c USP32-HA overexpression vs. vector control (Ctrl), n = 1 SILAC sample set. Small GTPases implicated in vesicular traffic whose modified peptides were detected are labeled according to their respective Log 2 ratios: magenta >1; green <−1; blue between −1 and 1, not significant. Analysis was performed using MaxQuant and Perseus software tools as described in the Methods under ubiquitome analysis. All mass spectrometry (MS) data can be accessed via the PRIDE repository (PXD011899). d Ubiquitylation status of GFP-Rab7 vs. GFP-Rab5 as a function of USP32 catalytic activity. GFP-Rabs, immunoprecipitated (IP) from HEK293T cells coexpressing HA-Ub and either USP32, C743A, or neither, was assessed by immunoblot against HA; WCL: whole cell lysate. See also Supplementary Fig. 5
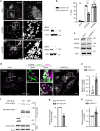
Fig. 5
USP32 regulates the Rab7-positive endosome. a Effect of USP32 depletion on Rab7-positive and Rab5-positive endosomes. Top panels: representative confocal images of endogenous Rab7 (white). Boxed region perinuclear (PN) and peripheral (PP) zoom overlays of Rab7 (green) highlight colocalization with late endosome (LE) marker LAMP1 (magenta). Bottom panels: representative confocal images of endogenous Rab5 (white). Boxed PN and PP region overlays of Rab5 (green) highlight colocalization with EE marker EEA1 (magenta). Cell and nuclear boundaries depicted in dashed magenta and white lines, respectively. Scale bars = 10 µm. b Rab pixel distribution expressed as fractional distance along a straight line from center of nucleus (0) to the PM (1.0). Red lines: mean, n = 2 independent experiments. c Alterations in LE morphology in response to USP32 depletion as visualized by correlative light and electron microscopy (CLEM). Overlays of GFP-Rab7 fluorescence (green) and transmission electron micrographs (TEM) are shown, scale bars = 0.25 µm. d Comparison of GFP-Rab7-positive LE profiles in control cells (siCtrl, black line) vs. those depleted of USP32 (siUSP32_2, red line), _x_-axis: LE diameter in µm, _y_-axis: number of LE profiles. See also Supplementary Fig. 6. e LE enlargement (white bars) and/or dispersion (gray bars) in MelJuSo cells depleted of the indicated GTPase (% cells), n = 4 or 5 independent experiments as indicated. See also Supplementary Fig. 7a. f Effects of USP32 depletion on cellular abundance of endogenous Rab7, as assessed by immunoblot. g Analysis of membrane-bound vs. cytosolic fractions of GFP-Rab7 stably expressed in MelJuSo cells with actin and transferrin receptor (TrfR) as loading controls for the cytosolic and membrane fractions, respectively. h Ratio of membrane-bound/cytosolic GFP-Rab7 in control cells (siCtrl, white bars) vs. those depleted of USP32 (siUSP32_2, gray bars) normalized to siCtrl, n = 3 independent experiments. i Schematic illustration of consequences of USP32 depletion on Rab7 membrane-to-cytosol equilibrium. Bar graphs report mean of independent measurements (black circles), error bars reflect ±s.d. Where applicable, total number of cells analyzed per condition appears above each bar/scatter. All significant values were calculated using Student’s t test: *p < 0.05, ***p < 0.001, NS = not significant
Fig. 6
Deubiquitylation of Rab7 by USP32 promotes late endosome transport. a Late endosome (LE) organization and dynamics as a function of Rab7 ubiquitylation status. Top panels: representative confocal images of live MelJuSo cells stably expressing GFP-Rab7 or GFP-Rab7-2KR (GFP-2KR) (white) taken at the start of time-lapse, t = 0. Scale bars = 10 µm. Bottom panels: vesicle displacement rates depicted on a rainbow color scale (blue: immobile; red: maximum mobility per time interval) tracked over 250 s at 5 s per frame. Cell and nuclear boundaries are depicted in dashed white lines, boxed zoom-ins highlight select perinuclear (PN) and peripheral (PP) regions. b Vesicle dispersion expressed as fractional distance of GFP pixels along a straight line from center of nucleus (0) to the PM (1.0). Red lines: mean, n = 2 independent experiments. c Quantification of vesicle motility calculated using TrackMate for Fiji (for details see the Methods section), n = 3 independent experiments. Plots report mean velocities of Lysotracker-positive structures calculated from multicell time-lapses of control cells (siCtrl, open circles) vs. those depleted of USP32 (siUSP32_2, closed circles). See also Supplementary Fig. 7b, c and Movies 9–12. d–f Co-immunoprecipitation (Co-IP) of Rab7-interacting protein (RILP) with GFP-Rab7 as a function of Rab7 ubiquitination status. d Immunoblots of Co-IP from HEK293T cells transfected and treated as indicated. e Quantification of Co-IP for HA-RILP with GFP-Rab7 (white bars) vs. GFP-2KR (gray bars), n = 3 independent experiments, is shown along with a schematic summary. f Quantification of Co-IP for HA-RILP with GFP-Rab7 from control HEK293T cells (siCtrl, white bars) vs. those depleted of USP32 (siUSP32_2, gray bars) normalized to control, n = 3 independent experiments. See also Supplementary Fig. 7d. Bar graphs report mean of independent measurements (black circles), error bars reflect ±s.d. Where applicable, total number of cells analyzed per condition appears above each bar/scatter. All significance was calculated using Student’s t test: *p < 0.05, **p < 0.01, ***p < 0.001, NS = not significant
Fig. 7
USP32 promotes retrograde trafficking by way of the retromer complex. a Effects of VPS35 and VPS26 depletion on the size and intracellular distribution of late endosomes (LEs). Representative confocal images of fixed MelJuSo cells transfected as indicated and immunostained against major histocompatibility class II (MHC-II) (white) are shown with the corresponding immunoblot analyses; targeting small interfering RNA (siRNA): (+); control siRNA: (−). Cell and nuclear boundaries are depicted in dashed magenta and white lines, respectively. b Percent cells harboring dispersed (gray bars) and/or enlarged (white bars) MHC-II-positive vesicles in response to VPS35 and VPS26 depletion, n = 3 independent experiments. c Effects of USP32 depletion on cellular abundance of endogenous VPS35 and VPS26, as assessed by immunoblot. d, e Effects of Rab7 ubiquitylation status on the retromer compartment. d Representative confocal images of fixed MelJuSo cells transferred with the indicated siRNAs and immunostained for endogenous VPS35 (white). Boxed perinuclear (PN) and peripheral (PP) region overlays of VPS35 (magenta) with MHC-II (green) highlight retromer/LE interactions. Cell and nuclear boundaries depicted in dashed magenta and white lines, respectively. e Colocalization between VPS35 and MHC-II in control cells (white bars) vs. those depleted of USP32 (gray bars). Plots report Mander’s overlap calculated from multicell images (black circles) taken from n = 3 independent experiments. See also Supplementary Fig. 8a–e. f–h Effect of Rab7 ubiquitylation status on its interaction with VPS35 as measured by proximity-based labeling with biotin. f Biotinylation of RFP-VPS35 in the presence of free GFP-BirA (−), GFP-BirA-Rab7 (WT) vs. GFP-BirA-2KR (2KR) assayed in HEK293T cells. g Biotinylation of RFP-VPS35 by GFP-BirA-Rab7 (white bar) vs. GFP-BirA-2KR (gray bar) above GFP-BirA background control, n = 3 independent experiments. h Quantification of endogenous VPS35 and RFP-VPS35 biotinylation (combined) by GFP-BirA-Rab7 above GFP-BirA control in control HeLa cells (siCtrl, white bar) vs. those depleted of USP32 (siUSP32_2, black bar), n = 5 independent experiments. Bar graphs report mean, error bars reflect ±s.d. See also Supplementary Fig. 8f. All significant values were calculated using Student’s t test: **p < 0.05, ***p < 0.001, NS = not significant. Sale bars = 10 µm
Fig. 8
USP32 regulates extraction of membranes from Rab7-positive endosomes. a, b Bud/tubule resolution from Rab7 endosomes as a function of USP32. Select confocal frame zooms taken from time-lapses of live control (siCtrl) vs. USP32-depleted (siUSP32_2) MelJuSo cells stably expressing (a) GFP-Rab7 or (b) GFP-2KR (green) labeled with Lysotracker (magenta) are shown. Large arrows point to emerging buds (B) and tubules (T), small arrows point to nascent vesicles formed as a result of fission (F). Quantification: number (#) of resolved (white) and unresolved (black) buds and tubules observed, n = 2 independent experiments. Scale bars = 5 µm. See also Supplementary Movies 13–16. c Alterations in late endosome (LE) morphology in response to USP32 depletion as visualized by correlative light and electron microscopy (CLEM). GFP-Rab7-2KR (GFP-2KR) fluorescence (green) and transmission electron micrographs (TEMs) are shown; scale bars = 0.25 µm. d Comparison of GFP-Rab7-2KR-positive LE profile in siCtrl (black line) and siUSP32_2 (red line); _x_-axis: LE diameter in µm; _y_-axis: number of LE profiles. See also Supplementary Fig. 9
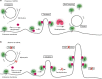
Fig. 9
Model of USP32 function in Rab7-mediated transport and recycling. a In the presence of USP32: non-ubiquitylated Rab7 can efficiently mediate minus end-directed transport, while C-terminal ubiquitination of Rab7 enables the switch to other functions, such as recycling from the late endosome/multi-vesicular body (LE/MVB). In turn, deubiquitylation of Rab7 by USP32 promotes fission of recycling tubules from the mother endosomes and release of Rab7 from the membrane (*refers to GDI (GDP Dissociation Inhibitor) associated with Rab7 upon release). b In the absence of USP32: lack of Rab7 deubiquitylation results in failure to resolve recycling tubules and inhibits liberation of Rab7 for subsequent functional cycles. (−): microtubule minus end, leading towards the perinuclear region; (+): microtubule plus end, leading towards the cell periphery
Similar articles
- Rab24 interacts with the Rab7/Rab interacting lysosomal protein complex to regulate endosomal degradation.
Amaya C, Militello RD, Calligaris SD, Colombo MI. Amaya C, et al. Traffic. 2016 Nov;17(11):1181-1196. doi: 10.1111/tra.12431. Epub 2016 Oct 3. Traffic. 2016. PMID: 27550070 - Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus.
Jaber N, Mohd-Naim N, Wang Z, DeLeon JL, Kim S, Zhong H, Sheshadri N, Dou Z, Edinger AL, Du G, Braga VM, Zong WX. Jaber N, et al. J Cell Sci. 2016 Dec 1;129(23):4424-4435. doi: 10.1242/jcs.192260. Epub 2016 Oct 28. J Cell Sci. 2016. PMID: 27793976 Free PMC article. - Dynein Is Required for Rab7-Dependent Endosome Maturation, Retrograde Dendritic Transport, and Degradation.
Yap CC, Digilio L, McMahon LP, Wang T, Winckler B. Yap CC, et al. J Neurosci. 2022 Jun 1;42(22):4415-4434. doi: 10.1523/JNEUROSCI.2530-21.2022. Epub 2022 Apr 26. J Neurosci. 2022. PMID: 35474277 Free PMC article. - Regulation of Endosomal Trafficking by Rab7 and Its Effectors in Neurons: Clues from Charcot-Marie-Tooth 2B Disease.
Mulligan RJ, Winckler B. Mulligan RJ, et al. Biomolecules. 2023 Sep 16;13(9):1399. doi: 10.3390/biom13091399. Biomolecules. 2023. PMID: 37759799 Free PMC article. Review. - Rab GTPase Function in Endosome and Lysosome Biogenesis.
Langemeyer L, Fröhlich F, Ungermann C. Langemeyer L, et al. Trends Cell Biol. 2018 Nov;28(11):957-970. doi: 10.1016/j.tcb.2018.06.007. Epub 2018 Jul 17. Trends Cell Biol. 2018. PMID: 30025982 Review.
Cited by
- Ubiquitomics: An Overview and Future.
Vere G, Kealy R, Kessler BM, Pinto-Fernandez A. Vere G, et al. Biomolecules. 2020 Oct 17;10(10):1453. doi: 10.3390/biom10101453. Biomolecules. 2020. PMID: 33080838 Free PMC article. Review. - Mechanisms regulating the sorting of soluble lysosomal proteins.
Meraş İ, Maes J, Lefrancois S. Meraş İ, et al. Biosci Rep. 2022 May 27;42(5):BSR20211856. doi: 10.1042/BSR20211856. Biosci Rep. 2022. PMID: 35394021 Free PMC article. Review. - EGFR-T790M Mutation-Derived Interactome Rerouted EGFR Translocation Contributing to Gefitinib Resistance in Non-Small Cell Lung Cancer.
Wu PS, Lin MH, Hsiao JC, Lin PY, Pan SH, Chen YJ. Wu PS, et al. Mol Cell Proteomics. 2023 Sep;22(9):100624. doi: 10.1016/j.mcpro.2023.100624. Epub 2023 Jul 24. Mol Cell Proteomics. 2023. PMID: 37495186 Free PMC article. - TRIM39 deficiency inhibits tumor progression and autophagic flux in colorectal cancer via suppressing the activity of Rab7.
Hu J, Ding X, Tian S, Chu Y, Liu Z, Li Y, Li X, Wang G, Wang L, Wang Z. Hu J, et al. Cell Death Dis. 2021 Apr 12;12(4):391. doi: 10.1038/s41419-021-03670-3. Cell Death Dis. 2021. PMID: 33846303 Free PMC article. - USP32 facilitates non-small cell lung cancer progression via deubiquitinating BAG3 and activating RAF-MEK-ERK signaling pathway.
Li S, Yang L, Ding X, Sun H, Dong X, Yang F, Wang M, Zhang H, Li Y, Li B, Liu C. Li S, et al. Oncogenesis. 2024 Jul 19;13(1):27. doi: 10.1038/s41389-024-00528-z. Oncogenesis. 2024. PMID: 39030175 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials