Cas9+ conditionally-immortalized macrophages as a tool for bacterial pathogenesis and beyond - PubMed (original) (raw)
Cas9+ conditionally-immortalized macrophages as a tool for bacterial pathogenesis and beyond
Allison W Roberts et al. Elife. 2019.
Abstract
Macrophages play critical roles in immunity, development, tissue repair, and cancer, but studies of their function have been hampered by poorly-differentiated tumor cell lines and genetically-intractable primary cells. Here we report a facile system for genome editing in non-transformed macrophages by differentiating ER-Hoxb8 myeloid progenitors from Cas9-expressing transgenic mice. These conditionally immortalized macrophages (CIMs) retain characteristics of primary macrophages derived from the bone marrow yet allow for easy genetic manipulation and a virtually unlimited supply of cells. We demonstrate the utility of this system for dissection of host genetics during intracellular bacterial infection using two important human pathogens: Listeria monocytogenes and Mycobacterium tuberculosis.
Keywords: Listeria monocytogenes; genome editing; host-pathogen interactions; immunology; infectious disease; inflammation; macrophage; microbiology; mouse; tuberculosis.
© 2019, Roberts et al.
Conflict of interest statement
AR, LP, GM, KC, DL, GG, GB, JC No competing interests declared
Figures
Figure 1.. Cas9+ CIMs as a tractable system for genome-editing in macrophages.
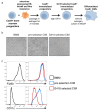
(a) Graphic overview of gene editing in Cas9+ CIMs. (b) BMMs (left panel), CIMs (middle), or RAW 264.7 cells (right) were visualized with Diff-Quick stain. (c) mRNA levels in BMMs and CIMs was quantified using a Nanostring nCounter. Data are representative of two independent experiments and are presented as log transformed normalized transcript counts of the average of technical duplicates from one experiment. (d) BMMs or CIMs transduced with a scramble guide (CIM scram) were analyzed by flow cytometry for expression of the indicated myeloid cell markers. Data are representative of three independent experiments. (e) IL-6 and IL-12p40 production by BMMs or CIM scram stimulated with the TLR4 ligand LPS or the TLR9 ligand CpG was measured by ELISA. N.D. – none detected. Data are representative of three independent experiments each performed in triplicate, mean ± SD are shown. (f) Genomic DNA from CIMs transduced with 40 guides targeting 17 genes was analyzed for genomic editing by TIDE analysis. (g) CIMs transduced with a scramble guide or guides targeting CD11b or Ifngr1 were stained with the indicated antibodies. Fluorescence minus one (FMO) stained samples were used as controls. (h and I) BMMs or CIMs transduced with a scramble guide or guides targeting Nos2 were stimulated overnight with LPS + IFNγ or left unstimulated and then analyzed by flow cytometry for expression of iNOS (h); nitric oxide production in cell-free supernatants was analyzed by Griess assay (i). Data are representative of two independent experiments each performed in triplicate, mean ± SD are shown. (j) CIM progenitors previously transduced with a lentivirus containing puromycin resistance and a guide targeting Ifngr1 were subsequently transduced with lentivirus containing hygromycin resistance and scramble guide or guides targeting CD11b.
Figure 1—figure supplement 1.. Re-selection of CIM progenitors that more closely resembled BMMs.
(a) Graphic overview of generation of Cas9+ CIMs. After generation of immortalized Cas9+ CIM progenitors, cells were re-selected with high concentrations of G418. (b) CIM morphology pre and post G418 selection. (c) BMMs or CIMs pre and post G418 selection were analyzed by flow cytometry for expression of the indicated myeloid cell markers.
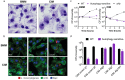
Figure 2.. Cas9+ CIMs as an in vitro model for Listeria monocytogenes infection.
(a) BMMs (left panel) or CIMs (right) were infected with WT Listeria monocytogenes at MOI = 0.25 and monolayers were visualized with Diff-Quick stain at 8 hr post-infection. Data are representative of two independent experiments. (b) BMMs (top three panels) or CIMs (lower panels) were infected for 5 hr with three strains of L. monocytogenes: WT, Δ_hly,_ and Δ_actA_ at MOI = 1.5. Nuclei shown in blue, bacterial cells in red, and F-actin in green. (c) BMMs (left panel) or CIMs transduced with a scramble gRNA were infected with three strains of L. monocytogenes: WT, an autophagy-sensitive strain that lacks ActA, PlcA and PlcB (Mitchell et al., 2018) and Δ_hly_ at MOI = 0.25. Bacterial densities were enumerated by CFU at t = 0.5 h and indicated hours post-infection. Data are representative of two independent experiments each performed in triplicate, mean ± SD are shown. (d) CIMs transduced with the indicated gRNAs were infected with WT L. monocytogenes or the autophagy-sensitive L. monocytogenes mutant at MOI = 0.75 and bacterial density was enumerated by CFU at t = 8 hr. Data are representative of two independent experiments each performed in triplicate, mean ± SD are shown.
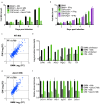
Figure 3.. Cas9+ CIMs as an in vitro model for Mycobacterium tuberculosis infection.
(a) Luminescent bacterial growth assay. BMMs (black) vs. CIMs transduced with a scramble guide (green) were infected with M. tuberculosis_-Erdman (solid bars) or a Δ_eccC M. tuberculosis_-Erdman strain (patterned bars) carrying the luxCDABE reporter operon at MOI = 0.5. Data are representative of two independent experiments each performed in triplicate, mean ± SD are shown. (b) Luminescent bacterial growth assay as in a, BMMs (black), CIMs transduced with a scramble guide (green), or CIMs transduced with a guide targeting Ifngr (purple) were infected with M. tuberculosis_-Erdman Lux. Patterned bars indicate conditions where the different cell types were treated with IFNγ prior to and throughout infection. Fold-change in luminescence is reported as M. tuberculosis growth relative to t = 0 for each condition. Data are representative of four independent experiments each performed in triplicate, mean ± SD are shown. (c–f) mRNA levels in BMMs and CIMs, either uninfected or infected with WT (c) or Δ_eccC M. tuberculosis (d) at MOI = 5 at 6 hr post infection were quantified using a Nanostring nCounter. Data are representative of two independent experiments. Source data is available as Figure 3—source data 1. c and (d) Data are presented as fold changes of infected/uninfected values of the average of technical duplicates from one experiment. (e) Log-transformed, normalized transcript counts for the indicated genes obtained from BMMs (black) vs. CIMs (green) before (patterned bars) or after infection with WT M. tuberculosis (solid bars). (f) Transcript counts for the indicated genes obtained from BMMs (black) vs. CIMs (green) infected with WT M. tuberculosis (solid bars) or M. tuberculosis Δ_eccC (patterned bars). Transcripts were normalized to counts in WT M. tuberculosis infected macrophages.
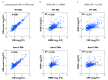
Figure 3—figure supplement 1.. Weak correlation between gene induction by RAW 264.7 cells and BMMs or CIMs in response to M. tuberculosis.
mRNA levels in BMMs, CIMs, or RAW 264.7 cells either uninfected or infected with WT or Δ_eccC M. tuberculosis_ at MOI = 5 at 6 hr post infection were quantified using a Nanostring nCounter. Data are from one experiment and are presented as fold changes of infected/uninfected values. Gene induction was compared between (a) untransduced CIMs vs CIMs transduced with a scramble gRNA (b) BMMs and RAW 264.7 cells and (C) CIMs and RAW 264.7 cells. Source data is available as Figure 3—source data 1.
Similar articles
- CRISPR/Cas9 Immunoengineering of Hoxb8-Immortalized Progenitor Cells for Revealing CCR7-Mediated Dendritic Cell Signaling and Migration Mechanisms in vivo.
Hammerschmidt SI, Werth K, Rothe M, Galla M, Permanyer M, Patzer GE, Bubke A, Frenk DN, Selich A, Lange L, Schambach A, Bošnjak B, Förster R. Hammerschmidt SI, et al. Front Immunol. 2018 Aug 28;9:1949. doi: 10.3389/fimmu.2018.01949. eCollection 2018. Front Immunol. 2018. PMID: 30210501 Free PMC article. - Ipr1 gene mediates innate immunity to tuberculosis.
Pan H, Yan BS, Rojas M, Shebzukhov YV, Zhou H, Kobzik L, Higgins DE, Daly MJ, Bloom BR, Kramnik I. Pan H, et al. Nature. 2005 Apr 7;434(7034):767-72. doi: 10.1038/nature03419. Nature. 2005. PMID: 15815631 Free PMC article. - Genetic modification of ER-Hoxb8 osteoclast precursors using CRISPR/Cas9 as a novel way to allow studies on osteoclast biology.
Di Ceglie I, van den Akker GG, Ascone G, Ten Harkel B, Häcker H, van de Loo FA, Koenders MI, van der Kraan PM, de Vries TJ, Vogl T, Roth J, van Lent PL. Di Ceglie I, et al. J Leukoc Biol. 2017 Apr;101(4):957-966. doi: 10.1189/jlb.1AB0416-180RR. Epub 2016 Dec 5. J Leukoc Biol. 2017. PMID: 27920208 - Cellular Microbiology: The metabolic interface between host cell and pathogen.
Russell DG. Russell DG. Cell Microbiol. 2019 Nov;21(11):e13075. doi: 10.1111/cmi.13075. Epub 2019 Jul 7. Cell Microbiol. 2019. PMID: 31231972 Free PMC article. Review. - Interactions of Listeria monocytogenes with the autophagy system of host cells.
Lam GY, Czuczman MA, Higgins DE, Brumell JH. Lam GY, et al. Adv Immunol. 2012;113:7-18. doi: 10.1016/B978-0-12-394590-7.00008-7. Adv Immunol. 2012. PMID: 22244576 Review.
Cited by
- History of tuberculosis disease is associated with genetic regulatory variation in Peruvians.
Nieto-Caballero VE, Reijneveld JF, Ruvalcaba A, Innocenzi G, Abeydeera N, Asgari S, Lopez K, Iwany SK, Luo Y, Nathan A, Fernandez-Salinas D, Chiñas M, Huang CC, Zhang Z, León SR, Calderon RI, Lecca L, Budzik JM, Murray M, Van Rhijn I, Raychaudhuri S, Moody DB, Suliman S, Gutierrez-Arcelus M. Nieto-Caballero VE, et al. PLoS Genet. 2024 Jun 13;20(6):e1011313. doi: 10.1371/journal.pgen.1011313. eCollection 2024 Jun. PLoS Genet. 2024. PMID: 38870230 Free PMC article. - Optimized Nonviral Gene Disruption in Primary Murine and Human Myeloid Cells.
Freund EC, Haag SM, Haley B, Murthy A. Freund EC, et al. Methods Mol Biol. 2023;2618:201-217. doi: 10.1007/978-1-0716-2938-3_15. Methods Mol Biol. 2023. PMID: 36905519 - Phagocytic cooperativity by tumour macrophages.
Maoz A, Weiskopf K. Maoz A, et al. Nat Biomed Eng. 2023 Sep;7(9):1057-1059. doi: 10.1038/s41551-023-01088-0. Nat Biomed Eng. 2023. PMID: 37679572 No abstract available. - Legionella pneumophila exploits the endo-lysosomal network for phagosome biogenesis by co-opting SUMOylated Rab7.
Li C, Fu J, Shao S, Luo ZQ. Li C, et al. PLoS Pathog. 2024 May 13;20(5):e1011783. doi: 10.1371/journal.ppat.1011783. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38739652 Free PMC article. - Particle uptake by macrophages triggers bifurcated transcriptional pathways that differentially regulate inflammation and lysosomal gene expression.
Cobo I, Murillo-Saich J, Alishala M, Calderon S, Coras R, Hemming B, Inkum F, Rosas F, Takei R, Spann N, Prohaska TA, Alabarse PVG, Jeong SJ, Nickl CK, Cheng A, Li B, Vogel A, Weichhart T, Fuster JJ, Le T, Bradstreet TR, Webber AM, Edelson BT, Razani B, Ebert BL, Taneja R, Terkeltaub R, Bryan RL, Guma M, Glass CK. Cobo I, et al. Immunity. 2025 Apr 8;58(4):826-842.e8. doi: 10.1016/j.immuni.2025.02.023. Epub 2025 Mar 20. Immunity. 2025. PMID: 40118070
References
- Chen M, Divangahi M, Gan H, Shin DS, Hong S, Lee DM, Serhan CN, Behar SM, Remold HG. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. The Journal of Experimental Medicine. 2008;205:2791–2801. doi: 10.1084/jem.20080767. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Open Philanthropy Fellow/Life Sciences Research Foundation/International
- U19 AI135990/AI/NIAID NIH HHS/United States
- R01AI072429/NH/NIH HHS/United States
- P01AI063302/NH/NIH HHS/United States
- U19AI135990/NH/NIH HHS/United States
- DP1AI24619/NH/NIH HHS/United States
- DP1 AI124619/AI/NIAID NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- R01 AI072429/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials