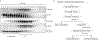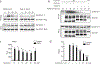UNC93B1 recruits syntenin-1 to dampen TLR7 signalling and prevent autoimmunity - PubMed (original) (raw)
UNC93B1 recruits syntenin-1 to dampen TLR7 signalling and prevent autoimmunity
Olivia Majer et al. Nature. 2019 Nov.
Abstract
At least two members of the Toll-like receptor (TLR) family, TLR7 and TLR9, can recognize self-RNA and self-DNA, respectively. Despite the structural and functional similarities between these receptors, their contributions to autoimmune diseases such as systemic lupus erythematosus can differ. For example, TLR7 and TLR9 have opposing effects in mouse models of systemic lupus erythematosus-disease is exacerbated in TLR9-deficient mice but attenuated in TLR7-deficient mice1. However, the mechanisms of negative regulation that differentiate between TLR7 and TLR9 are unknown. Here we report a function for the TLR trafficking chaperone UNC93B1 that specifically limits signalling of TLR7, but not TLR9, and prevents TLR7-dependent autoimmunity in mice. Mutations in UNC93B1 that lead to enhanced TLR7 signalling also disrupt binding of UNC93B1 to syntenin-1, which has been implicated in the biogenesis of exosomes2. Both UNC93B1 and TLR7 can be detected in exosomes, suggesting that recruitment of syntenin-1 by UNC93B1 facilitates the sorting of TLR7 into intralumenal vesicles of multivesicular bodies, which terminates signalling. Binding of syntenin-1 requires phosphorylation of UNC93B1 and provides a mechanism for dynamic regulation of TLR7 activation and signalling. Thus, UNC93B1 not only enables the proper trafficking of nucleic acid-sensing TLRs, but also sets the activation threshold of potentially self-reactive TLR7.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
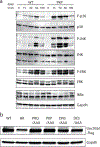
Extended Data Fig. 1:. A C-terminal region in Unc93b1 regulates TLR7 responses.
(a) Unc93b1PKP-expressing macrophages show enhanced TLR7 signaling. Immunoblot of P-p38, P-JNK, P-ERK, and IκBα of RAW macrophages stimulated with R848 (50 ng/ml) for indicated times. Representative of two independent experiments. (b) Unc93b1-Flag expression levels, as measured by Flag immunoblot, of Unc93b1-deficient RAW macrophages retrovirally transduced to express the indicated Unc93b1 alleles. These same cell lines are used for experiments shown in Fig. 1a. All data are representative of three independent experiments, unless otherwise noted.
Extended Data Fig. 2:. Unc93b1PKP does not alter TLR9 responses, unlike Unc93b1D34A.
(a) Representative flow cytometry analysis showing percent TNFα positive cells, measured by intracellular cytokine staining, of indicated RAW macrophage lines after stimulation with CpG-B (25 nM), R848 (10 ng/ml), ssRNA40 (1 µg/ml), PolyIC (20 µg/ml), or LPS (10 ng/ml). Shaded histograms show unstimulated controls. (b) TNFα production, measured by ELISA, from the indicated RAW macrophage lines after stimulation for 8h with R848 (10 ng/ml), CpG-B (25 nM), or LPS (50 ng/ml). Bars show mean of n=4 biological replicates pooled from two independent experiments (unpaired two-tailed Student’s t-test). (c) TLR7 and TLR9 trafficking are normal in Unc93b1PKP but not in Unc93b1D34A RAW lines. Immunoblot of TLR7-HA and TLR9-HA from lysates of indicated RAW macrophage lines; FL: full-length. All data are representative of three independent experiments.
Extended Data Fig. 3:. Unc93b1PKP does not alter TLR7 trafficking or localization.
(a) Unc93b1PKP does not alter TLR7 export rates. Pulse-chase analysis of TLR7 in Unc93b1WT and Unc93b1PKP-expressing RAW macrophages. Cell lysate was HA immunoprecipitated and subjected to radiolabeled screen and immunoblot. The full-length (FL) and cleaved forms of TLR7 are indicated. An asterisk denotes a nonspecific band. Representative of two independent experiments. (b) Unc93b1PKP does not affect TLR7 trafficking to endosomes. Levels of TLR7, Lamp1, and Calnexin in whole cell lysates (WCL) or lysates of purified phagosomes from the indicated RAW macrophage lines were measured by immunoblot. Representative of three independent experiments. (c) Colocalization of TLR7-HA and Lamp1 in RAW macrophages expressing the indicated Unc93b1-Flag alleles on a Myd88 −/− background using superresolution structured illumination microscopy. Shown are representative Unc93b1WT, Unc93b1PKP and Unc93b1H412R cells: TLR7 (red) and Lamp1 (green). Boxed areas are magnified. The plot shows quantification of the percentage of total TLR7 within Lamp1+ endosomes with each dot representing an individual cell. Data are pooled from two independent experiments. Scale bars: 10 µm. Bars show mean ± s.d. (unpaired two-tailed Student’s t-test). P-values are indicated. (d) The subcellular localization of Unc93b1PKP is not altered relative to Unc93b1WT. Colocalization of Unc93b1-Flag (red) and Lamp1 (green) was measured using superresolution structured illumination microscopy in Unc93b1-deficient RAW macrophages complemented with Unc93b1WT, Unc93b1PKP, or Unc93b1H412R. A representative cell is shown for each Unc93b1 allele. Boxed areas are magnified. The plot shows quantification of the percentage of total Unc93b1 within Lamp1+ endosomes with each dot representing an individual cell. Data acquired in a single experiment. Scale bars: 10µm. Bars show mean ± s.d. (unpaired two-tailed Student’s t-test). P-values are indicated.
Extended Data Fig. 4:. Mass spectrometry analysis of Unc93b1 complexes.
(a) A small fraction of Unc93b1 resides in endosomes compared to the endoplasmic reticulum (ER). Subcellular fractionation of TLR7-HA, Unc93b1-Flag expressing RAW macrophages was performed by density-gradient centrifugation. The distribution of Calnexin (ER), Lamp1 (late endosomes and lysosomes), Unc93b1-Flag, and TLR7-HA across fractions was measured by immunoblot. Representative of three independent experiments. (b) Workflow for isolation of phagosomes from RAW macrophages and purification of Unc93b1-Flag complexes from phagosome lysates.
Extended Data Fig. 5:. Syntenin-1 and Syntenin-2 inhibit TLR7 signaling.
(a) Syntenin-1 is selectively recruited to Unc93b1 upon TLR7 stimulation, but not TLR3 stimulation. Syntenin-1 binding to Unc93b1 was measured by Flag immunoprecipitation followed by immunoblot for Syntenin-1 from RAW macrophage lines stimulated with R848 (0.5 μg/ml) or PolyIC (10 µg/ml) for the indicated times. Levels of Syntenin-1 and Unc93b1-Flag in cell lysates are also shown. (b) Syntenin-1 associates selectively with the TLR7-Unc93b1 complex, but not with TLR9. Syntenin-1 binding to TLR7-HA or TLR9-HA was measured by HA immunoprecipitation followed by immunoblot for Syntenin-1 from indicated RAW macrophage lines stimulated with R848 (0.5 μg/ml) or CpG-B (0.5 µM) for the indicated times. Levels of Syntenin-1 and TLR7/9-HA in cell lysates are also shown. (c) NF-κB activation in HEK293T cells transiently expressing Syntenin-1 and stimulated with TNFα (10 ng/ml). (d) NF-κB activation in HEK293T cells transiently expressing TLR7 and increasing amounts of Syntenin-2. Cells were stimulated with R848 (50 ng/ml) for 16h prior to harvest. Data in c,d were measured using a dual luciferase reporter assay, normalized to Renilla expression and expressed as relative luciferase units (RLU). Bars show mean ± s.d., n=3 biological replicates, one-way ANOVA followed by a Tukey’s post test (95% confidence interval): *p < 0.05, **p < 0.01, ***p < 0.001. All data are representative of at least three independent experiments.
Extended Data Fig. 6:. Unc93b1K333R confers enhanced TLR7 signaling without affecting TLR9 and TLR3.
Flow cytometry analysis showing percent TNFα positive cells, measured by intracellular cytokine staining, of Unc93b1-deficient RAW macrophages expressing the indicated alleles after stimulation with CpG (25 nM), R848 (8 ng/ml), PolyIC (20 µg/ml), or LPS (10 ng/ml). Shaded histograms show unstimulated controls. Data are representative of three independent experiments.
Extended Data Fig. 7:. Serine phosphorylation in the C-terminal tail of Unc93b1 restricts TLR7 signaling.
(a,b) Validation of the anti-phospho-Unc93b1 polyclonal Unc93b1 antibody. (a) Immunoblots demonstrating the specificity of the phospho-specific antibodies generated against Ser547 and Ser550 in the Unc93b1 C-tail. Varying quantities of synthesized peptides corresponding to the Unc93b1 C-terminal regulatory region with (P-Unc93b1-C) and without (NP-Unc93b1-C) phosphorylated Ser547 and Ser550 were dropped onto membrane and probed with rabbit phospho-specific, affinity-purified polyclonal anti-Unc93b1 IgG. Representative of two independent experiments. (b) Phospho-specific polyclonal antibodies detect both phosphorylated Ser547 and Ser550. Unc93b1 was isolated from Unc93b1-deficient RAW macrophages expressing Unc93b1S547A, Unc93b1S550A, or Unc93b1S547A/S550A by Flag immunoprecipitation followed by immunoblot with phospho-specific polyclonal antibodies. Representative of at least three independent experiments. (c) Intracellular cytokine staining of TNFα in macrophage lines expressing the indicated Unc93b1 alleles and stimulated with CpG (10 nM), R848 (10 ng/ml) ssRNA40 (1 µg/ml), PolyIC (20 µg/ml), or LPS (10 ng/ml). Gray histograms are unstimulated controls. (d) TNFα production, measured by ELISA, from the indicated RAW macrophage lines after stimulation for 8 h with LPS (50 ng/ml). Bars show mean ± s.d., n=3 biological replicates. A representative of three independent experiments is shown. (e) Levels of phospho-p38 and phospho-JNK, as measured by immunoblot, in lysates of the indicated RAW macrophage cells stimulated with R848 (50 ng/ml). Representative of two independent experiments.
Extended Data Fig. 8:. Genetic variation in the human Unc93b1 C-terminal regulatory region increases TLR7 responses.
NFκB activation in HEK293T cells transiently expressing TLR7 or TLR5 and the indicated human Unc93b1 alleles was measured using a dual luciferase reporter assay. Cells were stimulated with R848 (10 ng/ml) or Flagellin (2 ng/ml) for 16h prior to harvest. Data are normalized to Renilla expression and expressed as relative luciferase units (RLUs). Bars show mean ± s.d., n=3 biological replicates; one-way ANOVA followed by a Tukey’s posttest (95% confidence interval): *p < 0.05, **p < 0.01, ***p < 0.001. Representative of three independent experiments.
Extended Data Fig. 9:. Unc93b1PKP knock-in mice develop systemic inflammation.
(a) CRISPR/Cas9 strategy to generate Unc93b1PKP knock-in mice. Green line indicates the guide sequence. Red bases indicate the edited codons. A representative sequencing trace of genomic DNA from an edited founder mouse is shown. (b) Flow cytometry analysis of the indicated immune cell populations in 6–8 weeks old Unc93b1 WT/WT, Unc93b1 PKP/WT, and Unc93b1 PKP/PKP mice. Frequencies of dendritic cells (CD11b+CD11c+MHCIIhigh) and inflammatory monocytes (CD11b+Ly6c+Ly6Gneg) in lymph nodes are shown. Data points were pooled from four independent experiments. P-values determined by unpaired two-tailed Student’s t-test are listed. (c) Unc93b1 PKP/PKP mice exhibit signs of emergency granulopoesis in their bone marrow compartment. Flow cytometry analysis of bone marrow from 6–8 week old Unc93b1 WT/WT, Unc93b1 PKP/WT, and Unc93b1 PKP/PKP mice. Gates representing LSK (CD45+CD3εnegCD19negLy6cnegLy6GnegSca-1highc-Kithigh) and Sca-1highc-Kitneg cells (CD45+CD3εnegCD19negLy6cnegLy6GnegSca-1highc-Kitneg) are indicated and compiled frequencies of Sca-1highc-Kitneg cells are shown on the right. Bars show mean ± s.d., n=3 biological replicates (_p_-value by unpaired two-tailed Student’s t-test). (d) Representative staining, corresponding to compiled results shown in Fig. 4c, of anti-nuclear antibodies (ANA) using sera from the indicated mouse ages and genotypes. (e,f) Flow cytometry analysis showing percent TNFα positive cells, measured by intracellular cytokine staining, of BMMs and BM-DCs derived from the indicated mice after stimulation with CpG-B (150 nM), R848 (10 ng/ml), ssRNA40 (1 µg/ml), PolyIC (10 µg/ml), or LPS (10 ng/ml). Shaded histograms are unstimulated controls. (g) TNFα production by BM-DCs derived from the indicated mice after stimulation for 8h with R848, CpG-B (150 nM), or LPS (50 ng/ml). (h) TNFα production by BMMs from the indicated mice after stimulation for 8h with CpG-B (500 nM), LPS (50 ng/ml), or increasing concentrations of R848. Data in g,h are mean ± s.d., n=3 biological replicates (_p_-values determined by unpaired two-tailed Student’s t-test). (i) B cells from Unc93b1 PKP/PKP mice show enhanced proliferation in response to TLR7 stimulation. Proliferation of CFSE-labeled B cells after 3 days stimulation with R848 (8 ng/ml) or LPS (1.6µg/ml) was measured by FACS, pre-gating on live CD19+ cells and quantifying the geometric mean fluorescent intensity (gMFI) of CSFE. The proliferation index is defined as gMFI CSFEUnstim: gMFI CFSESample. Bars are mean ± s.d., n=5 mice per group pooled together from three independent experiments (_p_-values determined by unpaired two-tailed Student’s t-test). (j) Immunoprecipitation of Myd88 from bone marrow-derived macrophages from the indicated mice after stimulation with R848 (500 ng/ml), followed by immunoblot for IRAK2. Input levels of Myd88 and IRAK2 in whole cell lysates (WCL) are also shown. (k) Unc93b1 protein levels in BMMs from indicated mouse genotypes, measured by immunoblot with polyclonal antibodies against endogenous Unc93b1. All data are representative of three independent experiments, unless otherwise noted.
Extended Data Fig. 10:. Gating strategies.
Representative gating strategies for marginal zone (MZ) B cells, activated T cells, dendritic cells, inflammatory monocytes, emergency granulopoesis in bone marrow, and B cell proliferation in splenocyte cultures are shown. These strategies were used for the data presented in Fig. 4 and Extended Data Fig. 9.
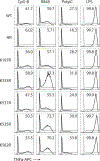
Fig. 1:. Syntenin-1 binds to the C-terminal tail of Unc93b1 and restricts TLR7 signaling.
(a) Intracellular cytokine staining of TNFα in macrophage lines expressing the indicated Unc93b1 alleles and stimulated with CpG-B (100 nM), R848 (10 ng/ml), ssRNA40 (2.5 µg/ml), PolyIC (20 µg/ml), or LPS (10 ng/ml). Shaded histograms are unstimulated controls. HR: Unc93b1H412R. (b) TNFα production, measured by ELISA, from the indicated RAW macrophage lines after stimulation for 8h with R848 (10 ng/ml), ssRNA40 (1 µg/ml), CpG-B (25 nM), or LPS (50 ng/ml). Data are mean ± s.d., n=2 biological replicates. PKP: Unc93b1PKP/AAA. (c) Topology of Unc93b1 with the C-terminal regulatory region indicated in orange. (d) Immunoprecipitation (IP) of MyD88 from RAW macrophage lines expressing the indicated Unc93b1-Flag alleles and stimulated with R848 (500 ng/ml) followed by immunoblot for IRAK2. Input levels of Myd88 and IRAK2 in whole cell lysates (WCL) are also shown. (e) Silver stained SDS-PAGE gel of purified Unc93b1-Flag complexes from phagosomes of RAW macrophages expressing the indicated Unc93b1-Flag alleles. The 32kD protein corresponding to Syntenin-1 is indicated. (f) Purified Unc93b1-Flag complexes described in (e) were immunoblotted for Syntenin-1. (g) Syntenin-1 binding to Unc93b1 was measured by Flag IP followed by immunoblot for Syntenin-1 from the indicated RAW macrophage lines stimulated with R848 (0.5 μg/ml). (h) Interaction between Syntenin-1 and Unc93b1 was measured as described in (g) from Unc93b1WT RAW macrophages stimulated with R848 (0.5 μg/ml) or CpG-B (0.5 μM). (i) NFκB activation in HEK293T cells transiently expressing TLR7 and increasing amounts of Syntenin-1 was measured using a dual luciferase reporter assay. Cells were stimulated with R848 (50 ng/ml) for 16h prior to harvest. RLUs: relative luciferase units, normalized to Renilla expression (n=3 biological replicates). One-way ANOVA, ***p<0.0001. All data are representative of at least three independent experiments.
Fig. 2:. Unc93b1-TLR7 complexes are sorted into intralumenal vesicles of multivesicular bodies.
(a) Exosome preparations are enriched for the exosome markers CD63, Alix, and Syntenin-1, and devoid of the ER marker Calnexin. Whole cell lysates (WCL) or lysates of exosomes from Unc93b1WT and Unc93b1HR-expressing RAW macrophages were probed with antibodies against the indicated proteins. Equivalent amounts of total protein were loaded per lane. (b) Sorting of TLR7 into exosomes of RAW macrophages requires Unc93b1. Immunoblot for TLR7-HA and Unc93b1-Flag of exosome preparations from Unc93b1WT and Unc93b1HR-expressing RAW macrophages. Samples were normalized to CD63 levels to ensure loading of similar exosome equivalents. (c) Unc93b1-Flag was immunoprecipitated from lysates of the indicated RAW macrophage lines and total or K63-linked ubiquitylation was measured by immunoblot. Arrowheads indicate the mobility of unmodified Unc93b1. (d) Schematic showing the relative positions of lysines analyzed in (c,e). (e) TNFα production, measured by ELISA, from the indicated RAW macrophage lines after stimulation for 8h with R848 (10 ng/ml), or LPS (10 ng/ml). Bars show mean of n=3 (n=2 for K197R and HR) biological replicates, representative of two independent experiments (_p_-value determined by unpaired two-tailed Student’s t-test). All other data are representative of at least three independent experiments.
Fig. 3:. Serine phosphorylation in the C-terminal tail of Unc93b1 regulates Syntenin-1 recruitment.
(a) TNFα production, measured by ELISA, from the indicated RAW macrophage lines after stimulation for 8h with R848 (20 ng/ml). Bars show mean ± s.d., n=3 biological replicates, _p_=0.0002 for PKP, p<0.0001 for all other mutants compared to WT (unpaired two-tailed Student’s t-test). (b) Syntenin-1 binding to Unc93b1WT or Unc93b1S547A/S550A in RAW macrophages stimulated with R848 (0.5 μg/ml) was measured by Unc93b1-Flag immunoprecipitation followed by immunoblot for Syntenin-1. (c) Unc93b1-Flag was immunoprecipitated from lysates of the indicated RAW macrophage lines and phosphorylation of Ser547 and Ser550 was measured by immunoblot with phospho-specific Unc93b1 antibodies. Each blot was performed on the same membrane but cropped to present relevant lanes. (d) RAW macrophages were stimulated with R848 (0.5 μg/ml) or CpG-B (0.5 µM) and Unc93b1 phosphorylation was measured by immunoblot after Unc93b1-Flag immunoprecipitation. (e) A model of Syntenin-1 recruitment to Unc93b1. All data are representative of at least three independent experiments.
Fig. 4:. Unc93b1PKP knock-in mice develop TLR7-driven systemic inflammation and autoimmunity.
(a) Gross appearance and weights of Unc93b1 PKP/PKP mice compared to littermate controls. (b) Flow cytometric analysis of the indicated immune cell populations in spleen or lymph nodes in Unc93b1 WT/WT, Unc93b1 PKP/WT, and Unc93b1 PKP/PKP mice at 6–8 weeks of age. Data points were pooled from four independent experiments. (c) Tabulated results showing the number and percentage of the indicated mice with anti-nuclear antibodies in their sera as measured by staining of Hep-2 slides. (d) Cells from Unc93b1 PKP/WT and Unc93b1 PKP/PKP mice show enhanced TLR7 responses. IL-12p40, IL-6, and IFN-I production, measured by ELISA, CBA, or bioassay, by bone marrow-derived dendritic cells (BM-DCs) derived from the indicated mice after stimulation for 8h with R848 (25 ng/ml or as indicated), CpG-B (150 nM), or LPS (50 ng/ml). Bars show mean ± s.d., n=3 biological replicates, representative of three independent experiments. (e) TLR7 deficiency rescues disease in Unc93b1 PKP/PKP mice. Weights and frequencies of the indicated cell populations from 6-week-old mice with indicated genotypes are shown. Data were pooled together from three independent experiments. (f) Proposed model of Syntenin-1-mediated restriction of TLR7 signaling. Upon activation of TLR7, Syntenin-1 binding to Unc93b1 may facilitate sorting of the TLR7/Unc93b1 complex into intraluminal vesicles (ILVs) of multivesicular bodies (MVBs), which terminates TLR7 signaling. Disruption of Syntenin-1 binding (e.g., in Unc93b1PKP expressing cells) prevents the inducible sorting of TLR7/Unc93b1 into MVBs, leading to unrestrained signaling and TLR7-driven autoimmunity. Additional mechanisms responsible for the steady-state turnover of TLR7 and/or Unc93b1 are not depicted. All _p_-values determined by unpaired two-tailed Student’s t-test.
Similar articles
- Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing.
Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, Beutler B, Miyake K. Fukui R, et al. J Exp Med. 2009 Jun 8;206(6):1339-50. doi: 10.1084/jem.20082316. Epub 2009 May 18. J Exp Med. 2009. PMID: 19451267 Free PMC article. - UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes.
Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. Kim YM, et al. Nature. 2008 Mar 13;452(7184):234-8. doi: 10.1038/nature06726. Epub 2008 Feb 27. Nature. 2008. PMID: 18305481 - Release from UNC93B1 reinforces the compartmentalized activation of select TLRs.
Majer O, Liu B, Woo BJ, Kreuk LSM, Van Dis E, Barton GM. Majer O, et al. Nature. 2019 Nov;575(7782):371-374. doi: 10.1038/s41586-019-1611-7. Epub 2019 Sep 23. Nature. 2019. PMID: 31546247 Free PMC article. - Regulatory molecules required for nucleotide-sensing Toll-like receptors.
Saitoh S, Miyake K. Saitoh S, et al. Immunol Rev. 2009 Jan;227(1):32-43. doi: 10.1111/j.1600-065X.2008.00729.x. Immunol Rev. 2009. PMID: 19120473 Review. - TLR7 Signaling in Lupus B Cells: New Insights into Synergizing Factors and Downstream Signals.
Satterthwaite AB. Satterthwaite AB. Curr Rheumatol Rep. 2021 Nov 24;23(11):80. doi: 10.1007/s11926-021-01047-1. Curr Rheumatol Rep. 2021. PMID: 34817709 Free PMC article. Review.
Cited by
- Identification of the Association Between Toll-Like Receptors and T-Cell Activation in Takayasu's Arteritis.
Tian Y, Huang B, Li J, Tian X, Zeng X. Tian Y, et al. Front Immunol. 2022 Jan 20;12:792901. doi: 10.3389/fimmu.2021.792901. eCollection 2021. Front Immunol. 2022. PMID: 35126357 Free PMC article. - Human Plasmacytoid Dendritic Cells and Cutaneous Melanoma.
Monti M, Consoli F, Vescovi R, Bugatti M, Vermi W. Monti M, et al. Cells. 2020 Feb 11;9(2):417. doi: 10.3390/cells9020417. Cells. 2020. PMID: 32054102 Free PMC article. Review. - Signaling Through Nucleic Acid Sensors and Their Roles in Inflammatory Diseases.
Okude H, Ori D, Kawai T. Okude H, et al. Front Immunol. 2021 Jan 28;11:625833. doi: 10.3389/fimmu.2020.625833. eCollection 2020. Front Immunol. 2021. PMID: 33633744 Free PMC article. Review. - Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer.
Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Han QF, et al. Mol Cancer. 2022 Nov 1;21(1):207. doi: 10.1186/s12943-022-01671-0. Mol Cancer. 2022. PMID: 36320056 Free PMC article. Review. - Targeting the innate immune receptor TLR8 using small-molecule agents.
Sakaniwa K, Shimizu T. Sakaniwa K, et al. Acta Crystallogr D Struct Biol. 2020 Jul 1;76(Pt 7):621-629. doi: 10.1107/S2059798320006518. Epub 2020 Jun 17. Acta Crystallogr D Struct Biol. 2020. PMID: 32627735 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- S10 OD018136/OD/NIH HHS/United States
- R01 AI105184/AI/NIAID NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- S10 RR025622/RR/NCRR NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- R01 AI072429/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials