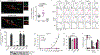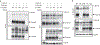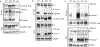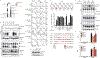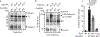Release from UNC93B1 reinforces the compartmentalized activation of select TLRs - PubMed (original) (raw)
Release from UNC93B1 reinforces the compartmentalized activation of select TLRs
Olivia Majer et al. Nature. 2019 Nov.
Abstract
Nucleic acid-sensing Toll-like receptors (TLRs) are subject to complex regulation to facilitate the recognition of microbial DNA and RNA while limiting the recognition of an organism's own nucleic acids1. Failure to properly regulate these TLRs can lead to autoimmune and autoinflammatory diseases2-6. Intracellular localization of these receptors is thought to be crucial for the discrimination between self and non-self7, but the molecular mechanisms that reinforce compartmentalized activation of intracellular TLRs remain poorly understood. Here we describe a mechanism that prevents the activation of TLR9 from locations other than endosomes. This control is achieved through the regulated release of the receptor from its trafficking chaperone UNC93B1, which occurs only within endosomes and is required for ligand binding and signal transduction. Preventing release of TLR9 from UNC93B1, either by mutations in UNC93B1 that increase affinity for TLR9 or through an artificial tether that impairs release, results in defective signalling. Whereas TLR9 and TLR3 are released from UNC93B1, TLR7 does not dissociate from UNC93B1 in endosomes and is regulated by distinct mechanisms. This work defines a checkpoint that reinforces the compartmentalized activation of TLR9, and provides a mechanism by which activation of individual endosomal TLRs may be distinctly regulated.
Conflict of interest statement
Author Information: The authors declare no competing financial interest. Readers are welcome to comment on the online version of the paper.
Figures
Extended Data Fig. 1:. A luminal Unc93b1 mutation results in defective TLR9 signaling despite normal trafficking.
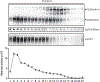
(a) Colocalization of Unc93b1 and Lamp1 in macrophages expressing the indicated Unc93b1 alleles using superresolution structured illumination microscopy. Shown are representative images: Unc93b1 (red) and Lamp1 (green). Boxed areas are magnified. The plot shows quantification of the percentage of total Unc93b1 within Lamp1+ endosomes. Each dot represents an individual cell. p values determined by unpaired two-tailed Student’s t-test. Data are from a single experiment. Scale bars: 10µm. (b) Unc93b1S282A is sufficient for the TLR9 signaling defect. NF-κB luciferase assay in HEK293T cells stimulated with CpG-B (1µM) for 16h. Data are normalized to Unc93b1-independent hIL-1b responses and expressed as luciferase fold change over unstimulated controls. n=3 biological replicates. *** indicates p<0.0001, ns = not significant (determined by unpaired two-tailed Student’s t-test). Blot below shows Unc93b1 expression levels. Representative of two independent experiments. (c) Intracellular cytokine staining of TNFα in macrophage lines expressing the indicated Unc93b1 alleles after stimulation with CpG-B (1 µM), R848 (100 ng/ml), PolyIC (100 ng/ml), Sa19 (200 ng/ml), Flagellin (100 ng/ml), and LPS (10 ng/ml). Gray histograms show unstimulated controls. Representative of two independent experiments. (d,e) TNFα production of the indicated macrophage lines after 8h stimulation with increasing concentrations of CpG-A (d), or LPS (50ng/ml) (e). n=2 biological replicates; *** indicates p<0.0001, ns = not significant, p values determined by two-way ANOVA followed by a Tukey’s posttest in (d) or by one-way ANOVA followed by a Tukey’s posttest in (e). Representative of two independent experiments. (f) Quantitative RT-PCR analysis of Tnfa expression in the indicated macrophage lines 8h after stimulation with CpG-A/DOTAP (1µM) or LPS (10ng/ml). n=3 biological replicates, *** indicates _p_=0.0003 for S282A vs WT) and _p_=0.0002 for HR vs WT, determined by unpaired two-tailed Student’s t-test. Representative experiment of two independent repeats. All data are shown as mean ± s.d.
Extended Data Fig. 2:. Unc93b1S282A does not affect DNA delivery to TLR9-containing endosomes.
(a) Uptake of Cy3-labeled CpG-B (1µM) of macrophage lines expressing the indicated alleles of Unc93b1-Flag. Data are presented as mean ± s.d. relative uptake compared to WT at 60 min. n=3 biological replicates. _p_-values determined by two-way ANOVA followed by a Tukey’s posttest when comparing WT vs SKN, ns = not significant. (b) Colocalization of CpG-A, TLR9-HA, and Lamp1 in macrophage lines shown in (a) after incubation with Cy3-labeled CpG-A (1µM) for 2h using superresolution structured illumination microscopy. Shown are representative images: TLR9 (green), Lamp1 (magenta), and CpG-A (red). Boxed areas and areas containing white lines are magnified. The histograms display fluorescent intensity plots of pixels along the white lines. Shaded areas highlight regions of colocalization of CpG-B, TLR9, and Lamp1. (c) Quantification of the percentage of CpG-A colocalized with TLR9. Each dot represents an individual cell, n=4 (WT), n=5 (SKN) and n=5 (HR). Data are mean ± s.d., *** indicates p<0.0001, ns = not significant. p values determined by unpaired two-tailed Student’s t-test. Scale bars: 5µm. Data are from a single experiment.
Extended Data Fig. 3:. Unc93b1S282A does not affect TLR9 dimerization or the association between N-terminal and C-terminal cleavage products of TLR9.
(a) Macrophage lines co-expressing TLR9-HA and TLR9-V5 together with the indicated Unc93b1-Flag alleles were subjected to HA-immunoprecipitation followed by V5-immunoblot. TLR9 levels in whole cell lysates (WCL) are also shown. (b) Macrophage lines expressing TLR9-HA and the indicated Unc93b1-Flag alleles were subjected to immunoprecipitation with an anti-TLR9 antibody specific to the N-terminal cleavage fragment (B33A4), followed by immunoblot of the C-terminal TLR9 fragment with anti-HA. Data are representative of at least two independent experiments.
Extended Data Fig. 4:. Unc93b1S282A shows a stronger interaction with TLR9, but not TLR7.
(a) Unc93b1SKN and Unc93b1S282A display a stronger association with TLR9. Immunoprecipitation of TLR9-HA from macrophage lines expressing the indicated Unc93b1 alleles, followed by immunoblot of Unc93b1-Flag. (b) Unc93b1SKN and Unc93b1S282A do not affect the interaction with TLR7. Immunoprecipitation of Unc93b1-Flag from macrophage lines expressing TLR7-HA and the indicated Unc93b1 alleles, followed by immunoblot of TLR7-HA. (c) Immunoprecipitation of Unc93b1-Flag from macrophage lines expressing the indicated Unc93b1 alleles, followed by immunoblot of TLR9-HA. All blots are representative of at least two independent experiments. SA: S282A. KA: K283A. NA: N284A. WCL: whole cell lysate. FL: full-length.
Extended Data Fig. 5:. Identification of residues within loop 5 of Unc93b1 that mediate interaction with TLR9.
(a) Schematic of the tested loop 5 mutants of Unc93b1 and the relative TLR9 responses indicated in shades of gray: white indicates a response equivalent to WT while black indicates no response. Asterisks show human Unc93b1 SNPs that have been tested in (f). (b) A larger region in loop 5 of Unc93b1 mediates binding to TLR9. Immunoprecipitation of Unc93b1-Flag from macrophage lines expressing the indicated Unc93b1 mutants (spanning amino acids 267–284, and non-functional HR) followed by immunoblot of TLR9-HA. Representative of two independent experiments. (c) Intracellular cytokine staining of TNFα in macrophage lines shown in (b) after stimulation with CpG-B (25 nM), R848 (100 ng/ml), and LPS (10 ng/ml). Shaded histograms show unstimulated controls. Representative of three independent experiments. (d) Schematics showing relative positions and a sequence alignment (bottom) of swapped regions within the TLR9/3 chimeras. Colored regions indicate TLR3 sequences. NF-κB luciferase assay in HEK293T cells transiently transfected with the indicated TLR9 and Unc93b1 mutants and stimulated with CpG-B (200nM) for 16h. Data are normalized to Unc93b1-independent hIL-1b responses and expressed as luciferase fold change over unstimulated controls. Data are mean ± s.d., n=3 biological replicates. _p_-values determined by two-way ANOVA followed by a Sidak’s posttest comparing each TLR9 allele coexpressed with Unc93b1 WT vs S282A (TLR9WT: p<0.0001, TLR9Mut1: _p_>0.9999, TLR9Mut2: p<0.0001, TLR9Mut3: p<0.0001, TLR9Mut4: _p_=0.0020, TLR9Mut5: _p_=0.0171. Representative experiment of two independent repeats. (e) TLR9 mutants that rescue signaling in the presence of Unc93b1S282A also show normal binding to Unc93b1S282A. HA immunoprecipitation of the indicated TLR9 mutants transiently expressed in HEK293T cells that stably express the indicated Unc93b1-Flag alleles, followed by immunoblot of Unc93b1-Flag. Representative of three independent experiments. (f) Human Unc93b1 variants with SNPs in loop 5 show decreased TLR9 signaling. NF-κB luciferase assay in HEK293T cells expressing TLR9 or TLR7 and the indicated human Unc93b1-Flag variants and stimulated with CpG-B (250 nM) or R848 (250 ng/ml) for 16h, respectively. Data are normalized to Renilla expression and expressed as relative luciferase units (RLUs). Data are mean ± s.d., n=3 biological replicates. _p_-values are determined by one-way ANOVA followed by a Tukey’s posttest. For CpG-B stimulations: _p_=0.0048 (WT vs G270S), _p_=0.0113 (WT vs R277Q), _p_=0.9994 (WT vs G283R). Representative of four independent experiments. For R848 stimulations: _p_=0.2001 (WT vs G270S), _p_=0.0002 (WT vs R277Q), _p_=0.9933 (WT vs G283R). Representative of three independent repeats experiment. (g) Expression levels of the Unc93b1 mutants used in (f). *p< 0.05, **p< 0.01, ***p< 0.001, ns: not significant. WCL: whole cell lysate. FL: full-length.
Extended Data Fig. 6:. Cellular fractionation showing the distribution profiles for CpG-B-biotin ligand and β-hexosaminidase.
Macrophages were stimulated for 4h with biotinylated CpG-B (1µM) and subjected to sub-cellular fractionation by density-gradient centrifugation. The distributions of TLR9-HA, CpG-B, Lamp1, and β-hexosaminidase activity are shown. Representative of two independent experiments.
Extended Data Fig. 7:. The TLR9-Unc93b1 association is reduced in endosomes compared to the ER.
(a) Input controls of TLR9 and Unc93b1 (relates to Fig. 3b). TLR9 and Unc93b1 levels in pooled ER or endosome fractions from macrophage lines expressing TLR9-HA and the indicated Unc93b1 alleles. FL: full-length. Representative of three independent experiments. (b) Increased interaction between TLR9 and Unc93b1S282A in endosomes. Immunoprecipitation of Unc93b1-Flag from pooled endosome fractions followed by immunoblot for TLR9-HA. Input controls are also shown. Representative of three independent experiments. Bar graph shows the quantification of TLR9 bound to Unc93b1 in pooled endosome fractions, normalized by Unc93b1-Flag levels in endosome fractions. Data are mean ± s.d., each dot represents data from an independent experiment (n=3). * indicates _p_=0.0413 (unpaired two-tailed Student’s t-test). (c) Release model of TLR9.
Extended Data Fig. 8:. Tethering of TLR9 and Unc93b1.
(a) Cysteine mutants of TLR9 and Unc93b1 do not affect trafficking of TLR9 to endosomes. Immunoblot of TLR9-HA from macrophage lines expressing the indicated TLR9-HA and Unc93b1-Flag cysteine mutants. Representative of two independent experiments. (b) TLR9-HA immunoblot under non-reducing conditions after immunoprecipitation of TLR9-HA from macrophage lines shown in (a). The high molecular weight band indicates disulfide-bond formation between Unc93b1 and TLR9. Representative of two independent experiments. (c) Unc93b1-tethered TLR9 is unable to signal. NF-κB luciferase assay in HEK293T cells expressing the indicated cysteine mutant combinations and stimulated with CpG-B (1µM) for 16h. Data are normalized to Renilla expression and expressed as luciferase fold change over unstimulated controls. Data are mean ± s.d., n=3 biological replicates. _p_-values determined by unpaired two-tailed Student’s t-test. Representative of three independent experiments. FL: full-length.
Extended Data Fig. 9:. TLR3 but not TLR7 releases from Unc93b1 in endosomes.
(a) Subcellular fractionation of macrophages lines showing the distributions of TLR3, TLR7, and TLR9 across fractions. The pooled endosome and ER fractions for subsequent coimmunoprecipitations are highlighted. (b) Immunoprecipitation of TLR9- and TLR3-HA from pooled ER or endosome fractions of macrophage lines expressing wildtype Unc93b1. Immunoprecipitated TLR-HA levels were normalized across fractions and probed for levels of Unc93b1-Flag. Bar graph shows the quantification of Unc93b1 bound to TLR3 between ER and endosome fractions. Data are mean ± s.d., each dot represents data from an independent experiment (n=3). ** indicates _p_=0.0039 (paired two-tailed Student’s t-test). (c) Input controls of TLR9, TLR7 and Unc93b1 in pooled ER and endosome fractions (relates to Fig. 3d). (d) Less TLR3 or TLR9 is associated with Unc93b1WT in pooled endosome fractions compared to TLR7. Immunoprecipitation of Unc93b1-Flag from pooled endosome fractions (as shown in (a)) followed by immunoblot for TLR3-, TLR7-, or TLR9-HA. Bar graph shows the calculated relative proportion of Unc93b1-bound TLR compared to total amount of the same TLR in the pooled endosome fractions. Data represent the mean ± s.d., each dot represents data from an independent experiment (n=3). ** indicates _p_=0.0032 (TLR3 vs TLR7) and _p_=0.0036 (TLR7 vs TLR9), determined by unpaired two-tailed Student’s t-test. All immunoblots are representative of three independent experiments. FL: full-length.
Extended Data Fig. 10.. Generation of Unc93b1S282A knock-in mice and B-cell stimulation with LPS.
(a) CRISPR/Cas-9 strategy to generate Unc93b1S282A knock-in mice. Blue line indicates the guide sequence. Red bases indicated the edited codon. A representative sequencing trace of genomic DNA from an edited founder mouse is shown. (b) B cell proliferation in CSFE-labeled splenocyte cultures of the indicated mouse genotypes after stimulation for 3 days with increasing doses of LPS. The proliferation index is defined as geometric fluorescent intensity (gMFI) CSFEUnstim: gMFI CFSESample. Each curve shows the dose response of cells from three mice. Data are mean ± s.d. p values determined by two-way ANOVA followed by a Sidak’s posttest, ns = not significant. (c) Gating strategy for B cell stimulation assay.
Fig. 1:. An Unc93b1 mutation results in defective TLR9 signaling despite normal trafficking.
(a) Schematic of Unc93b1 topology with the SKN/AAA mutation indicated. (b) Intracellular cytokine staining of TNFα in Unc93b1-deficient RAW macrophage lines expressing the indicated Unc93b1 alleles (WT, SKN, and non-functional HR) after stimulation with CpG-B or CpG-A (150nM), R848 (25ng/ml), PolyIC (500ng/ml), or LPS (10ng/ml). Shaded histograms show unstimulated controls. (c) Immunoblot of TLR9-HA from the RAW macrophage lines shown in (b). FL: full-length. (d) TNFα production of the indicated RAW macrophage lines after 8h stimulation with increasing concentrations of CpG. n=2 biological replicates, representative of two independent experiments. (e) Quantitative RT-PCR analysis of Ifnb expression in the indicated RAW macrophage lines after 8h stimulation with CpG-A/DOTAP (1µM); n=3 biological replicates, representative of two independent experiments. (f) Whole cell (WCL) or phagosome lysates of the indicated RAW macrophage lines probed for TLR9-HA, Lamp-1, Calnexin, and Gapdh. All data are mean ± s.d.; _p_-values in (d) determined by two-way ANOVA followed by a Tukey’s post-test, WT vs S282A: * indicates _p_=0.0208, *** indicates p<0.0001; _p_-values in (e) determined by unpaired two-tailed Student’s t-test, *** indicates p<0.0001. All data are representative of at least three independent experiments, unless otherwise noted.
Figure 2.. Unc93b1S282A attenuates ligand binding and increases association with TLR9.
(a) Biotin-CpG-bound TLR9 complexes were precipitated from lysates of RAW macrophage lines expressing TLR9-HA and the indicated Unc93b1 alleles after stimulation with Biotin-CpG (1µM) for 4h, followed by immunoblot for TLR9-HA. Bars show mean ± s.d. of CpG-bound TLR9 relative to total TLR9; n=3 (WT), n=4 (SKN or S282A), and n=2 (HR), each dot represents an independent experiment. (b) Immunoprecipitation of Unc93b1-Flag from the RAW macrophage lines in (a) followed by immunoblot of TLR9-HA. TLR9 levels in whole cell lysates (WCL) are also shown. Representative of six independent experiments. Bars show mean ± s.d. of TLR9 bound to Unc93b1; n=6 (WT) n=7 (SKN or S282A); each dot represents an independent experiment. (c) Proximity ligation assay to analyze interaction between TLR9-HA and the indicated Unc93b1 alleles in RAW macrophages. Representative images are shown. Control staining was performed by omitting primary anti-HA antibodies. Quantification of PLA signals per cell volume is shown below images. Each dot represents an individual cell. Data are pooled from two independent experiments. Scale bar = 10 µm (d) Immunoprecipitation of TLR9-HA from phagosome preparations of RAW macrophage lines in (a) followed by immunoblot for Unc93b1-Flag. TLR9 and Unc93b1 levels in phagosomes and whole cell lysates are also shown. Representative of two independent experiments. All _p_-values determined by unpaired two-tailed Student’s t-test.
Fig. 3:. TLR9, but not TLR7, must release from Unc93b1 for signaling.
(a) Subcellular fractionation of RAW macrophage lines showing the distributions of TLR9, Unc93b1, and the indicated organelle markers. The pooled fractions, enriched for endosomes or ER, used for subsequent co-IP experiments are boxed. (b) Immunoprecipitation of TLR9-HA from pooled ER or endosome (Endo) fractions as shown in (a), followed by immunoblots for Unc93b1-Flag and TLR9-HA. Total TLR9-HA levels were normalized across fractions. Input controls are shown in Extended Data Fig. 7a. Bar graph shows mean ± s.d. of Unc93b1 bound to TLR9 across fractions. Each dot represents data from an independent experiment (n=3). _p_-values determined by paired two-tailed Student’s t-test. (c) Immunoprecipitation of TLR9-HA under non-reducing conditions from RAW macrophage lines expressing the indicated TLR9-HA and Unc93b1-Flag cysteine mutants, followed by immunoblot for Unc93b1. The non-reducing condition visualizes disulfide-bond formation between Unc93b1 and TLR9, which disappears after treatment with dithiothreitol (reducing condition). Bar graph below shows an NF-κB luciferase assay in HEK293T cells expressing the indicated cysteine mutant combinations and stimulated with CpG-B (1µM) for 16h. Data are normalized to Renilla expression and shown as relative luciferase units (RLUs); n=4 biological replicates. *** indicates p<0.0001 (unpaired two-tailed Student’s t-test.). (d) Immunoprecipitation of TLR9- and TLR7-HA from pooled ER or endosome fractions of macrophage lines co-expressing Unc93b1-Flag, followed by Flag and HA immunoblots. Input controls are shown in Extended Data Fig. 9c. Bar graph shows mean ± s.d. of Unc93b1 bound to TLR9 or TLR7 across ER and endosome fractions. Each dot represents data from an independent experiment (n=3). _p_-values determined by paired two-tailed Student’s t-test.
Fig. 4:. Release from Unc93b1 is required for TLR9 function in vivo.
(a-c) Immune cells from Unc93b1S282A mice show a selective loss of TLR9 signaling. (a) Intracellular cytokine staining of TNFα in bone marrow-derived macrophages from Unc93b1WT/- and Unc93b1S282A/- mice after stimulation with CpG-B (0.5µM), R848 (25 ng/ml), Poly IC (20µg/ml), or LPS (50ng/ml). Data are mean ± s.d.; n=3 biological replicates. *** indicates p<0.0001, ns = not significant (unpaired two-tailed Student’s t-test). Representative of three independent experiments. (b) IFN-I production by pDCs after 16h stimulation with the indicated concentrations of CpG-A. Each data point represents one mouse (n=3). Data are mean ± s.d.; _p_-values determined by two-way ANOVA followed by a Tukey’s post-test. For WT compared to S282A, ** indicates _p_=0.0064 and *** indicates p<0.0001. Representative of three independent experiments. (c) Proliferation of CFSE-labeled splenic B cells from the indicated mice after stimulation for 3d with CpG-B or R848. The proliferation index is defined as geometric fluorescent intensity (gMFI) CSFEUnstim: gMFI CFSESample. Each curve shows the dose response of cells from three separate mice. Data are mean ± s.d.; _p_-values determined by two-way ANOVA followed by a Sidak’s post-test. *** indicates p<0.0001; ns = not significant.
Comment in
- Regulation of endosomal TLRs.
Minton K. Minton K. Nat Rev Immunol. 2019 Nov;19(11):660-661. doi: 10.1038/s41577-019-0229-1. Nat Rev Immunol. 2019. PMID: 31562494 No abstract available.
Similar articles
- Cell Surface Expression of Endosomal Toll-Like Receptors-A Necessity or a Superfluous Duplication?
Mielcarska MB, Bossowska-Nowicka M, Toka FN. Mielcarska MB, et al. Front Immunol. 2021 Feb 1;11:620972. doi: 10.3389/fimmu.2020.620972. eCollection 2020. Front Immunol. 2021. PMID: 33597952 Free PMC article. Review. - UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes.
Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. Kim YM, et al. Nature. 2008 Mar 13;452(7184):234-8. doi: 10.1038/nature06726. Epub 2008 Feb 27. Nature. 2008. PMID: 18305481 - UNC93B1 and nucleic acid-sensing Toll-like receptors mediate host resistance to infection with Leishmania major.
Schamber-Reis BL, Petritus PM, Caetano BC, Martinez ER, Okuda K, Golenbock D, Scott P, Gazzinelli RT. Schamber-Reis BL, et al. J Biol Chem. 2013 Mar 8;288(10):7127-36. doi: 10.1074/jbc.M112.407684. Epub 2013 Jan 16. J Biol Chem. 2013. PMID: 23325805 Free PMC article. - Acidic amino acid residues in the juxtamembrane region of the nucleotide-sensing TLRs are important for UNC93B1 binding and signaling.
Kim J, Huh J, Hwang M, Kwon EH, Jung DJ, Brinkmann MM, Jang MH, Ploegh HL, Kim YM. Kim J, et al. J Immunol. 2013 May 15;190(10):5287-95. doi: 10.4049/jimmunol.1202767. Epub 2013 Apr 12. J Immunol. 2013. PMID: 23585677 - Regulatory molecules required for nucleotide-sensing Toll-like receptors.
Saitoh S, Miyake K. Saitoh S, et al. Immunol Rev. 2009 Jan;227(1):32-43. doi: 10.1111/j.1600-065X.2008.00729.x. Immunol Rev. 2009. PMID: 19120473 Review.
Cited by
- Profiling Differential Effects of 5 Selective Serotonin Reuptake Inhibitors on TLRs-Dependent and -Independent IL-6 Production in Immune Cells Identifies Fluoxetine as Preferred Anti-Inflammatory Drug Candidate.
Takenaka Y, Tanaka R, Kitabatake K, Kuramochi K, Aoki S, Tsukimoto M. Takenaka Y, et al. Front Pharmacol. 2022 Jun 22;13:874375. doi: 10.3389/fphar.2022.874375. eCollection 2022. Front Pharmacol. 2022. PMID: 35814203 Free PMC article. - Cell Surface Expression of Endosomal Toll-Like Receptors-A Necessity or a Superfluous Duplication?
Mielcarska MB, Bossowska-Nowicka M, Toka FN. Mielcarska MB, et al. Front Immunol. 2021 Feb 1;11:620972. doi: 10.3389/fimmu.2020.620972. eCollection 2020. Front Immunol. 2021. PMID: 33597952 Free PMC article. Review. - Use of Fluorescent Chemical Probes in the Study of Toll-like Receptors (TLRs) Trafficking.
Franco AR, Artusa V, Peri F. Franco AR, et al. Methods Mol Biol. 2023;2700:57-74. doi: 10.1007/978-1-0716-3366-3_3. Methods Mol Biol. 2023. PMID: 37603174 - Cyclical palmitoylation regulates TLR9 signalling and systemic autoimmunity in mice.
Ni H, Wang Y, Yao K, Wang L, Huang J, Xiao Y, Chen H, Liu B, Yang CY, Zhao J. Ni H, et al. Nat Commun. 2024 Jan 2;15(1):1. doi: 10.1038/s41467-023-43650-z. Nat Commun. 2024. PMID: 38169466 Free PMC article. - Aberrant B Cell Signaling in Autoimmune Diseases.
Corneth OBJ, Neys SFH, Hendriks RW. Corneth OBJ, et al. Cells. 2022 Oct 27;11(21):3391. doi: 10.3390/cells11213391. Cells. 2022. PMID: 36359789 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- S10 OD018136/OD/NIH HHS/United States
- R01 AI105184/AI/NIAID NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- R01 AI072429/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials