Identification of HCMV-derived T cell epitopes in seropositive individuals through viral deletion models - PubMed (original) (raw)
. 2020 Mar 2;217(3):jem.20191164.
doi: 10.1084/jem.20191164.
Stefanie Spalt 1 2, Daniel J Kowalewski 1, Cosima Zimmermann 3 4, Liane Bauersfeld 3 4, Annika Nelde 1 5, Leon Bichmann 1 6, Ana Marcu 1, Janet Kerstin Peper 1, Oliver Kohlbacher 6 7 8 9, Juliane S Walz 5, Vu Thuy Khanh Le-Trilling 10, Hartmut Hengel 3 4, Hans-Georg Rammensee 1 2, Stefan Stevanović 1 2, Anne Halenius 3 4
Affiliations
- PMID: 31869419
- PMCID: PMC7062530
- DOI: 10.1084/jem.20191164
Identification of HCMV-derived T cell epitopes in seropositive individuals through viral deletion models
Maren Lübke et al. J Exp Med. 2020.
Abstract
In healthy individuals, immune control of persistent human cytomegalovirus (HCMV) infection is effectively mediated by virus-specific CD4+ and CD8+ T cells. However, identifying the repertoire of T cell specificities for HCMV is hampered by the immense protein coding capacity of this betaherpesvirus. Here, we present a novel approach that employs HCMV deletion mutant viruses lacking HLA class I immunoevasins and allows direct identification of naturally presented HCMV-derived HLA ligands by mass spectrometry. We identified 368 unique HCMV-derived HLA class I ligands representing an unexpectedly broad panel of 123 HCMV antigens. Functional characterization revealed memory T cell responses in seropositive individuals for a substantial proportion (28%) of these novel peptides. Multiple HCMV-directed specificities in the memory T cell pool of single individuals indicate that physiologic anti-HCMV T cell responses are directed against a broad range of antigens. Thus, the unbiased identification of naturally presented viral epitopes enabled a comprehensive and systematic assessment of the physiological repertoire of anti-HCMV T cell specificities in seropositive individuals.
© 2019 Lübke et al.
Conflict of interest statement
Disclosures: M. Lübke reported a patent to HCMV epitopes (pending). S. Spalt reported a patent to HCMV epitopes (pending) and is currently an employee of Immatics Biotechnologies GmbH. D.J. Kowalewski reported a patent to HCMV epitopes (pending) and is currently an employee of Immatics Biotechnologies. C. Zimmermann reported a patent to HCMV epitopes (pending). L. Bauersfeld reported a patent to HCMV epitopes (pending). A. Nelde reported a patent to HCMV epitopes (pending). V.T.K. Le-Trilling reported a patent to HCMV epitopes (pending). H. Hengel reported a patent to HCMV epitopes (pending). H.-G. Rammensee reported "other" from Immatics GmbH and "other" from Curevac AG during the conduct of the study; in addition, H.-G. Rammensee had a patent to HCMV epitopes (pending). A. Halenius reported a patent to HCMV epitopes (pending). No other disclosures were reported.
Figures
Figure 1.
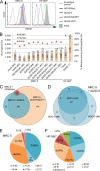
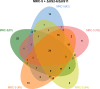
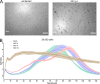
Deletion of the genes US2-11 allows identification of HCMV-derived HLA ligands from fibroblasts by LC-MS/MS. (A) MRC-5 and HF-99/7 fibroblasts were mock-treated or infected with AD169VarL wild-type virus or deletion mutants with an MOI of 5. Cell surface expression of HLA-I (W6/32) was analyzed by flow cytometry at 48 h.p.i. Shown are representative results of two independent experiments. (B) Overview of HLA ligand identifications obtained by LC-MS/MS analysis of MRC-5 cells after mock treatment (n = 2 independent experiments), infection with AD169VarL (n = 1), and infection with the deletion viruses AD169 ΔUS2-6 (n = 3) or ΔUS2-6/ΔUS11 (n = 5). Identified ligands of mock treated (n = 1) or AD169 ΔUS2-11 infected (n = 1) HF-99/7 cells are depicted on the right side. Cells were infected with an MOI of 4–7. Peptide identifications were defined as HLA ligands if they showed predicted HLA binding defined as NetMHC IC50 ≤ 500 nM and/or normalized SYFPEITHI scores ≥50%. The purity of HLA ligand extracts (i.e., the ratio of predicted binders/total peptide identifications) of the individual HLA ligand elutions is indicated by red triangles. (C) Overlap analysis of the combined datasets of HCMV-derived HLA ligands identified on MRC-5 cells infected with the three different virus variants. (D) Overlap of HCMV-derived HLA ligands identified in three independent experiments using MRC-5 cells infected with the deletion virus ΔUS2-6. (E and F) Distribution of HLA restrictions among the 198 (MRC-5; E) and 181 (HF-99/7; F) unique HCMV-derived HLA ligands identified in total. Ab, antibody; IDs, identifications.
Figure S1.
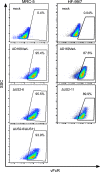
Flow cytometry–based determination of the infection rate. Cells were permeabilized and treated with Fc-FITC, which binds to the HCMV-encoded Fc-receptor (vFcR). Infection rate was determined once per infection experiment. SSC, side scatter.
Figure S2.
Overlap analysis of HCMV-derived HLA ligands between five independent HLA ligand elutions from MRC-5 cells infected with AD169 ΔUS2-6/ΔUS11.
Figure 2.
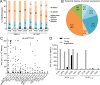
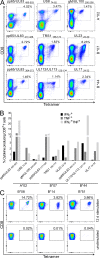
Identification and characterization of naturally presented T cell epitopes by ELISpot assays. HCMV ligands were tested for memory T cell response by IFNγ ELISpot with PBMCs of healthy, seropositive donors. (A) Distribution of dominant and subdominant HCMV ligands restricted to HLA-A and -B allotypes. (B) Proportion of epitope source proteins assigned to five different temporal classes of protein expression according to Weekes et al. (2014). Source proteins not assigned to one of those classes are depicted as not determined (ND). (C) ELISpot screening results of positively tested HLA-B*07:02-restricted peptides. Shown is the mean number of IFNγ spot-forming cells (SFCs) from two technical replicates for each tested donor minus the mean spot number of the negative control of the respective donor. Spot counts of >1,000 were set to 1,000 because of inaccurate spot count due to technical limitations. Positively evaluated spot counts are depicted in black, and negatively evaluated spot counts in gray. (D) Comparison of IFNγ SFC in ex vivo ELISpots (black) and ELISpots with prior 12-d amplification (gray). Shown are exemplary results of two independent experiments with five donors (coded by four-digit numbers) each for two B*08 epitopes (UL34180–188 and UL2661–69). ND, not determined.
Figure S3.
Results of ELISpot screening. (A–G) ELISpot screening of positively tested peptides with HLA-A*02:01 (A), HLA-A*29:02 (B), HLA-B*44:02 (C), HLA-A*01:01 (D), HLA-A*03:01 (E), HLA-B*08:01 (F), and HLA-B*51:01 (G) restriction. Depicted are numbers of IFNγ SFCs for each tested donor minus the spot numbers of the negative control of the respective donor. Positive evaluated donors are depicted in black, and negative tested donors in gray. Epitopes were tested once in 9–38 different HLA-matched donor PBMCs.
Figure S4.
Parallel recognition of multiple HCMV epitopes. (A and B) Exemplary ELISpot results after 12-d amplification with HLA-B*07-restricted (A) and HLA-B*44-restricted (B) epitopes using PBMCs of two and three donors, respectively. PBMCs were stimulated with 10 novel and already known epitopes (columns 1–10). (A) pp65/UL83265–275 (RPHERNGFTVL, column 10) is a previously identified epitope that was not contained in the ligands identified here. HIV Nef128–137 (TPGPGVRYPL) and medium served as negative controls, and PHA as positive control. Representative results of two independent experiments. (B) pp65/UL83364–373 (SEHPTFTSQY) and pp65/UL83511–521 (QEFFWDANDIY) are already known epitopes that were not found as ligands in this study. UL57193–203 (EEIPASDDVLF) served as negative control. Representative results of two independent experiments.
Figure S5.
Infection of MRC-5 cells with AD169 ΔUS2-6 for following cytotoxicity testing of peptide-specific T cell clones. (A) Comparison of morphology of uninfected and infected (20 h.p.i., MOI 1) MRC-5 cells. Scale bars, 200 µm. (B) Titration of MOIs in comparison with mock-treated MRC-5 cells for the xCELLigence system. 20,000 cells/well of infected or mock-treated MRC-5 cells were seeded into 96-well E-plates. Impedance was measured every 15 min and normalized to impedance of wells with medium only. The resulting dimensionless normalized cell index indicates the changes in impedance normalized to _t_0. Experiment was performed in triplicate. Shown are representative results of three independent experiments.
Figure 3.
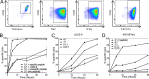
Characterization of HCMV-specific memory T cells. (A) Representative tetramer staining after 12-d in vitro amplification of CD8+ T cells derived from HLA-matched healthy donors. Novel HCMV ligands are compared with known pp65 epitopes (left column). (B) Exemplary intracellular IFNγ and TNF staining of healthy donor PBMCs after 12-d amplification measured by flow cytometry. Cells were stimulated with novel HCMV peptides or known pp65 epitopes. Bars represent percentage of CD8+ T cells producing IFNγ (black), TNF (gray), or both (light gray). Three peptides per HLA restriction, tested in one HLA-matched donor, are depicted. (C) Representative UL2661–69 (LPYPRGYTL)-tetramer staining after 12-d in vitro amplification or no stimulation of CD8+ T cells derived from HLA-matched healthy donors. Donor PBMCs (HLA-B*08+, B*14+, or B*51+) were initially stimulated with UL2661–69. Staining was performed with UL2661–69 tetramers specific for the respective HLA allotype. All tetramer stainings and ICS were performed once in two to four different donors per epitope.
Figure 4.
Characterization and cytotoxicity of HCMV-specific CD8+ T cell clones. (A) Exemplary staining of a UL2322–30-specific T cell clone (B*07:02-restricted) with the respective tetramer and ICS with TNF, IFNγ, and the degranulation marker CD107a. Similar results were obtained with several clones per donors. (B–D) Real-time cytotoxicity of different UL2322–30-specific T cell clones monitored by the xCELLigence system. 20,000 cells/well of infected or mock-treated MRC-5 cells were seeded into 96-well E-plates. After attachment of target cells, effector cells were added 48 h.p.i. at indicated E:T ratios (_t_0). Synthetic peptides were added to target cells 1 h before effector cells. Percentage of lysis was calculated in relation to cells without effector T cells. Shown are representative results of three independent experiments. Error bars represent SD from technical triplicates. (B) MRC-5 cells were loaded with specific (UL2322–30, RPWKPGQRV) or unspecific (HIV Nef128–137, TPGPGVRYPL) peptide or infected with ΔUS2-6 (MOI 2) and incubated with effector cells in an E:T cell ratio of 5:1. MRC-5 cells without peptide served as a control. (C) Comparison of specific lysis of ΔUS2-6–infected MRC-5 cells with different E:T ratios. (D) Specific lysis of AD169VarL wild-type infected or peptide-loaded cells with indicated E:T cell ratios.
Figure 5.
Position of 9mer ligands assigned to HLA-A*02:01 in the whole proteome prediction by SYFPEITHI against their immunogenicity. Dominant and subdominant epitopes are depicted in red and blue, respectively. Nonimmunogenic peptides are depicted in black. The dashed line indicates the best 2% of the prediction that should contain the majority of T cell epitopes according to SYFPEITHI. A possible correlation between the rank of HLA binding affinity predictions and the immunogenicity was assessed with the Kendall rank correlation coefficient tau-b (τb = −0.225, P = 0.033).
Similar articles
- Human Macrophages Escape Inhibition of Major Histocompatibility Complex-Dependent Antigen Presentation by Cytomegalovirus and Drive Proliferation and Activation of Memory CD4+ and CD8+ T Cells.
Frascaroli G, Lecher C, Varani S, Setz C, van der Merwe J, Brune W, Mertens T. Frascaroli G, et al. Front Immunol. 2018 May 25;9:1129. doi: 10.3389/fimmu.2018.01129. eCollection 2018. Front Immunol. 2018. PMID: 29887865 Free PMC article. - Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers.
Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, Khanna R. Elkington R, et al. J Virol. 2003 May;77(9):5226-40. doi: 10.1128/jvi.77.9.5226-5240.2003. J Virol. 2003. PMID: 12692225 Free PMC article. - Presentation of an immunodominant immediate-early CD8+ T cell epitope resists human cytomegalovirus immunoevasion.
Ameres S, Mautner J, Schlott F, Neuenhahn M, Busch DH, Plachter B, Moosmann A. Ameres S, et al. PLoS Pathog. 2013;9(5):e1003383. doi: 10.1371/journal.ppat.1003383. Epub 2013 May 23. PLoS Pathog. 2013. PMID: 23717207 Free PMC article. Clinical Trial. - Generation, maintenance and tissue distribution of T cell responses to human cytomegalovirus in lytic and latent infection.
Jackson SE, Sedikides GX, Okecha G, Wills MR. Jackson SE, et al. Med Microbiol Immunol. 2019 Aug;208(3-4):375-389. doi: 10.1007/s00430-019-00598-6. Epub 2019 Mar 20. Med Microbiol Immunol. 2019. PMID: 30895366 Free PMC article. Review. - Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells.
Vieira Braga FA, Hertoghs KM, van Lier RA, van Gisbergen KP. Vieira Braga FA, et al. Eur J Immunol. 2015 Sep;45(9):2433-45. doi: 10.1002/eji.201545495. Epub 2015 Aug 24. Eur J Immunol. 2015. PMID: 26228786 Review.
Cited by
- The T-cell response to SARS-CoV-2: kinetic and quantitative aspects and the case for their protective role.
Bertoletti A, Tan AT, Le Bert N. Bertoletti A, et al. Oxf Open Immunol. 2021 Feb 23;2(1):iqab006. doi: 10.1093/oxfimm/iqab006. eCollection 2021. Oxf Open Immunol. 2021. PMID: 38626271 Free PMC article. Review. - _Orf Virus_-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes.
Reguzova A, Ghosh M, Müller M, Rziha HJ, Amann R. Reguzova A, et al. Vaccines (Basel). 2020 Jun 10;8(2):295. doi: 10.3390/vaccines8020295. Vaccines (Basel). 2020. PMID: 32531997 Free PMC article. - SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition.
Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lübke M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Traenkle B, Kaiser PD, Rothbauer U, Becker M, Junker D, Krause G, Strengert M, Schneiderhan-Marra N, Templin MF, Joos TO, Kowalewski DJ, Stos-Zweifel V, Fehr M, Rabsteyn A, Mirakaj V, Karbach J, Jäger E, Graf M, Gruber LC, Rachfalski D, Preuß B, Hagelstein I, Märklin M, Bakchoul T, Gouttefangeas C, Kohlbacher O, Klein R, Stevanović S, Rammensee HG, Walz JS. Nelde A, et al. Nat Immunol. 2021 Jan;22(1):74-85. doi: 10.1038/s41590-020-00808-x. Epub 2020 Sep 30. Nat Immunol. 2021. PMID: 32999467 - Broad and Efficient Activation of Memory CD4+ T Cells by Novel HAdV- and HCMV-Derived Peptide Pools.
Höttler A, März L, Lübke M, Rammensee HG, Stevanović S. Höttler A, et al. Front Immunol. 2021 Jul 7;12:700438. doi: 10.3389/fimmu.2021.700438. eCollection 2021. Front Immunol. 2021. PMID: 34322126 Free PMC article. - HCMV Variants Expressing ULBP2 Enhance the Function of Human NK Cells via its Receptor NKG2D.
Meyer G, Siemes AR, Kühne JF, Bevzenko I, Baszczok V, Keil J, Beushausen K, Wagner K, Steinbrück L, Messerle M, Falk CS. Meyer G, et al. Eur J Immunol. 2025 Feb;55(2):e202451266. doi: 10.1002/eji.202451266. Eur J Immunol. 2025. PMID: 39931744 Free PMC article.
References
- Britten C.M., Gouttefangeas C., Welters M.J., Pawelec G., Koch S., Ottensmeier C., Mander A., Walter S., Paschen A., Müller-Berghaus J., et al. . 2008. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol. Immunother. 57:289–302. 10.1007/s00262-007-0378-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials