Abrogation of esophageal carcinoma development in miR-31 knockout rats - PubMed (original) (raw)
. 2020 Mar 17;117(11):6075-6085.
doi: 10.1073/pnas.1920333117. Epub 2020 Mar 2.
Cristian Taccioli 3, Alexey Palamarchuk 4, Guidantonio Malagoli Tagliazucchi 5, Ruiyan Jing 6, Karl J Smalley 2, Sili Fan 7, Joseph Altemus 8, Oliver Fiehn 7, Kay Huebner 4, John L Farber 6, Carlo M Croce 9
Affiliations
- PMID: 32123074
- PMCID: PMC7084137
- DOI: 10.1073/pnas.1920333117
Abrogation of esophageal carcinoma development in miR-31 knockout rats
Louise Y Fong et al. Proc Natl Acad Sci U S A. 2020.
Abstract
MicroRNA-31 (miR-31) is overexpressed in esophageal squamous cell carcinoma (ESCC), a deadly disease associated with dietary Zn deficiency and inflammation. In a Zn deficiency-promoted rat ESCC model with miR-31 up-regulation, cancer-associated inflammation, and a high ESCC burden following _N_-nitrosomethylbenzylamine (NMBA) exposure, systemic antimiR-31 delivery reduced ESCC incidence from 85 to 45% (P = 0.038) and miR-31 gene knockout abrogated development of ESCC (P = 1 × 10-6). Transcriptomics, genome sequencing, and metabolomics analyses in these Zn-deficient rats revealed the molecular basis of ESCC abrogation by miR-31 knockout. Our identification of EGLN3, a known negative regulator of nuclear factor κB (NF-κB), as a direct target of miR-31 establishes a functional link between oncomiR-31, tumor suppressor target EGLN3, and up-regulated NF-κB-controlled inflammation signaling. Interaction among oncogenic miR-31, EGLN3 down-regulation, and inflammation was also documented in human ESCCs. miR-31 deletion resulted in suppression of miR-31-associated EGLN3/NF-κB-controlled inflammatory pathways. ESCC-free, Zn-deficient miR-31-/- rat esophagus displayed no genome instability and limited metabolic activity changes vs. the pronounced mutational burden and ESCC-associated metabolic changes of Zn-deficient wild-type rats. These results provide conclusive evidence that miR-31 expression is necessary for ESCC development.
Keywords: constitutive miR-31 knockout rat; esophageal cancer rat model; esophageal squamous cell carcinoma; in vivo antimiR-31 delivery; zinc deficiency.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
Fig. 1.
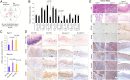
Systemic delivery of locked nucleic acid-mediated antimiR-31 suppresses esophageal carcinogenesis. (A) Study design. (B) Body weights, serum Zn levels, and esophageal miR-31 levels (qPCR) (rat snoRNA as normalizer; Tukey-HSD post hoc unpaired t test for multiple comparisons; error bars represent SD; n = 8 rats per group). (C) Macroscopic view of whole esophagus. Representative photos of ZD:CTRL (R13, R14) and ZD:CTRL-A/20-wk (R15, R16) with multiple large/sessile esophageal tumors vs. ZD:antimiR/20-wk (R1 to R4) esophagi with small/isolated tumors (arrowheads). (D) Tumor multiplicity (number of tumors per esophagus; error bars represent mean ± SD) (two-tailed Welch t test; n = 8 to 20 rats per cohort); large tumor (size > 2 mm) and ESCC incidence (%) (two-tailed Fisher’s exact test; n = 8 to 20 rats per cohort). (E) Histopathologic changes (arrowheads highlight the most relevant histological findings) in ZD esophagus after antimiR-31 treatment. Representative photos of ZD:antimiR/20-wk vs. ZD:CTRL esophagus showing H&E staining, IHC staining for KRT14 (brown, 3,3′-diaminobenzidine tetrahydrochloride; DAB), and PCNA (red, 3-amino-9-ethylcarbazole substrate chromogen; AEC) and ISH localization of miR-31 (blue, BCIP/NBT; counterstain, nuclear fast red) (n = 8 rats per cohort).
Fig. 2.
Genetic miR-31 knockout completely prevents ESCC development. (A) Study design. (B) Body weights, serum Zn levels, and qPCR analysis of esophageal miR-31 levels (snoRNA as normalizer; error bars represent mean ± SD; Welch’s t test; n = 12 rats per cohort; ns, not significant). (C) Macroscopic view of whole esophagus. Representative photos of ZD:miR-31−/− vs. ZD:miR-31+/+ esophagus (arrowheads indicate esophageal tumors). (D) Overall tumor incidence and ESCC incidence (%) (Fisher’s exact test) and tumor multiplicity (number of tumors per esophagus; error bars represent mean ± SD) (Welch t test). Statistical tests were two-sided (n = 20 rats per ZD cohort). (E) miR-31 localization in esophagus. Representative photos of ZD:miR-31−/− vs. ZD:miR-31+/+ tissues (miR-31 ISH signal; blue, BCIP/NBT) (n = 20 rats per cohort). Arrowheads indicate esophageal epithelium. (F) Histopathologic changes in miR-31−/− rat esophagus (arrowheads emphasize the difference in epithelia response to dietary ZD in miR-31−/− vs. miR-31+/+ esophagus and the similarity in the epithelia response in the ZS:miR-31−/− vs. ZS:miR-31+/+ esophagus). Representative photos of ZD:miR-31−/− vs. ZD:miR-31+/+ and ZS:miR-31−/− vs. ZS:miR-31+/+ esophagus showing histology (H&E), IHC staining for KRT14 (brown, DAB), and PCNA (red, AEC) (n = 20 rats per cohort). (G) Prevalence of somatic mutations in esophageal epithelia in ZD:CTRL (WT), ZS:CTRL (WT), and ZD:miR-31−/− rats with divergent esophageal tumor outcome. ZD:CTRL (R13) and ZD:CTRL (R14) esophagus, respectively, had high and low tumor burden; ZS:CTRL had no observable tumors, and nonproliferative ZD:miR-31−/− esophagus was tumor-free.
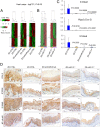
Fig. 3.
miR-31-5p targets EGLN3 and MBOAT2 expression. (A) Venn diagram showing the intersection between 86 genes that were the most derepressed after antimiR-31 treatment and predicted miR-31 target genes from TargetScan human/rat software. (B) Transfection experiments were performed in HEK-293FT cells using constructs for 3 predicted miR-31 targets, MBOAT2, EGLN3, and CTNND2. Renilla luciferase activity was normalized to firefly luciferase activity. The normalized luciferase activity in 293FT cells transfected with empty psiCHECK2 vector and scrambled negative control 1 (premiR-N.C.1) and relative luciferase activities of 293FT cells transfected with all other combinations of psiCHECK2 constructs and premiR oligos are shown. Experiments were repeated thrice in duplicate and data are presented as mean ± SD. Luciferase reporter assays indicate direct interactions between miR-31-5p and MBOAT2 (P < 0.005) and between miR-31-5p and EGLN3 (P < 0.001), two-tailed Student’s t test. (C) mRNA expression of Egln3 and Mboat2 in esophageal mucosa-derived RNA from ZD:CTRL, ZS:CTRL, and ZD:miR-31−/− rats (Psmb6 as normalizer; error bars represent mean ± SD; n = 7 to 11 rats). P values represent the significant difference between each individual group and ZD:CTRL rats; two-tailed, one-way ANOVA and Tukey post hoc t tests. (D) Protein expression of miR-31 targets EGLN3, MBOAT2, and STK40 (arrowheads emphasize the abundant/intense miR-31 overexpression in ZD:CTRL vs. ZS:CTRL esophagus). Representative photos of ZD:CTRL, ZS:CTRL, ZD:miR-31−/−, and ZS:miR-31−/− esophagus showing miR-31 ISH signal (blue, NBT/BCIP) (EGLN3, MBOAT2, and STK40 protein expression; brown, DAB; n = 10 rats per cohort). (E) Connection between miR-31 expression, protein expression of EGLN3 and STK40, and inflammation markers in human ESCC tissue. Representative photos of human ESCC tissue (FFPE sections): Case 1, case 2, and normal esophageal mucosa in case 2 showing miR-31 ISH signal (blue, NBT/BCIP) (EGLN3, STK40, COX-2, S100A8, S100A9, and NF-κB p65 protein expression; brown, DAB; n = 10 patient cases).
Fig. 4.
In vivo miR-31 silencing abolishes overexpression of cancer-related inflammation genes in esophagus. (A) Heatmaps (logFC 1, P < 0.05) showing distinct expression profiles of ZD:CTRL vs. ZS:CTRL, ZD:antimiR/5-wk vs. ZS:CTRL, and ZD:antimiR/20-wk vs. ZS:CTRL. (B) Heatmaps (logFC 1, P < 0.05) showing distinct expression profiles of ZD:miR-31−/− vs. ZS:CTRL and ZD:miR-31−/− vs. ZS:miR-31−/−. (C) mRNA expression (qPCR) of S100a8, Ptgs2, and S100a9 (top up-regulated inflammation genes;
SI Appendix, Table S1
) in ZD:CTRL, ZS:CTRL, ZD:antimiR/20-wk, and ZD:miR-31−/− esophagus (error bars represent mean ± SD; n = 7 to 11 rats per cohort; P values represent the significant difference between each individual group and the ZD:CTRL group; two-tailed, one-way ANOVA and Tukey post hoc t tests). (D) Protein expression (IHC) of COX-2, S100A8, S100A9, and NF-κB p65 (arrowheads emphasize the overexpression of the four inflammation markers in ZD:CTRL esophagus; brown, DAB; n = 10 rats per cohort). Representative photos of ZD:CTRL, ZS:CTRL, ZD:antimiR/20-wk, ZD:miR-31−/−, and ZS:miR-31−/− esophagus.
Fig. 5.
Metabolomics profiling by GC-TOF MS reveals divergent esophageal metabolic phenotypes in ZD:CTRL (WT) and ZD:miR-31−/− rats. (A) ESCC-bearing ZD:CTRL vs. ZS:CTRL esophagus showing 38 up- and 31 down-regulated metabolites. (B) Tumor-free ZD:miR-31−/− vs. ZS:miR-31−/− esophagus showing limited metabolic changes (eight up- and three down-regulated metabolites). Each node denotes an identified metabolite (red, up-regulated; blue, down-regulated; yellow, insignificant change; Mann–Whitney U test, P < 0.05). Metabolite size reflects median fold change. Metabolites are connected based on biochemical relationships (red lines) or structural similarity (blue lines). Molecules not directly participating in biochemical transformations but sharing structural properties were connected at a threshold of Tanimoto similarity coefficient ≥0.7 (n = 9 rats per group).
Similar articles
- Zinc treatment reverses and anti-Zn-regulated miRs suppress esophageal carcinomas in vivo.
Fong LY, Huebner K, Jing R, Smalley KJ, Brydges CR, Fiehn O, Farber JL, Croce CM. Fong LY, et al. Proc Natl Acad Sci U S A. 2023 May 16;120(20):e2220334120. doi: 10.1073/pnas.2220334120. Epub 2023 May 8. Proc Natl Acad Sci U S A. 2023. PMID: 37155893 Free PMC article. - Repression of Esophageal Neoplasia and Inflammatory Signaling by Anti-miR-31 Delivery In Vivo.
Taccioli C, Garofalo M, Chen H, Jiang Y, Tagliazucchi GM, Di Leva G, Alder H, Fadda P, Middleton J, Smalley KJ, Selmi T, Naidu S, Farber JL, Croce CM, Fong LY. Taccioli C, et al. J Natl Cancer Inst. 2015 Aug 18;107(11):djv220. doi: 10.1093/jnci/djv220. Print 2015 Nov. J Natl Cancer Inst. 2015. PMID: 26286729 Free PMC article. - MicroRNA dysregulation and esophageal cancer development depend on the extent of zinc dietary deficiency.
Fong LY, Taccioli C, Jing R, Smalley KJ, Alder H, Jiang Y, Fadda P, Farber JL, Croce CM. Fong LY, et al. Oncotarget. 2016 Mar 8;7(10):10723-38. doi: 10.18632/oncotarget.7561. Oncotarget. 2016. PMID: 26918602 Free PMC article. - Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals.
Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z. Huang H, et al. Cancer Biol Ther. 2019;20(1):73-80. doi: 10.1080/15384047.2018.1507254. Epub 2018 Sep 12. Cancer Biol Ther. 2019. PMID: 30207835 Free PMC article. - ZFPM2-AS1 facilitates cell growth in esophageal squamous cell carcinoma via up-regulating TRAF4.
Sun G, Wu C. Sun G, et al. Biosci Rep. 2020 Apr 30;40(4):BSR20194352. doi: 10.1042/BSR20194352. Biosci Rep. 2020. PMID: 32065218 Free PMC article.
Cited by
- Integrated analyses reveal IDO1 as a prognostic biomarker coexpressed with PD-1 on tumor-associated macrophages in esophageal squamous cell carcinoma.
Peng Y, Wang L, Yang J, Wu Q, Sun X, Zhang J, Yu Y, Zhang L, Gao J, Zhou Q, Zhu H, Yin F. Peng Y, et al. Front Pharmacol. 2024 Sep 16;15:1466779. doi: 10.3389/fphar.2024.1466779. eCollection 2024. Front Pharmacol. 2024. PMID: 39351094 Free PMC article. - Micronutrients Importance in Cancer Prevention-Minerals.
Saeed RF, Awan UA, Aslam S, Qazi AS, Bhatti MZ, Akhtar N. Saeed RF, et al. Cancer Treat Res. 2024;191:145-161. doi: 10.1007/978-3-031-55622-7_6. Cancer Treat Res. 2024. PMID: 39133407 Review. - Circulating long non-coding RNA EWSAT1 acts as a liquid biopsy marker for esophageal squamous cell carcinoma: A pilot study.
Uttam V, Rana MK, Sharma U, Singh K, Jain A. Uttam V, et al. Noncoding RNA Res. 2023 Oct 28;9(1):1-11. doi: 10.1016/j.ncrna.2023.10.009. eCollection 2024 Mar. Noncoding RNA Res. 2023. PMID: 38028735 Free PMC article. - MiR-199a-5p Decreases Esophageal Cancer Cell Proliferation Partially through Repression of Jun-B.
Phatak P, Tulapurkar ME, Burrows WM, Donahue JM. Phatak P, et al. Cancers (Basel). 2023 Sep 30;15(19):4811. doi: 10.3390/cancers15194811. Cancers (Basel). 2023. PMID: 37835506 Free PMC article. - circFNDC3B promotes esophageal squamous cell carcinoma progression by targeting MYO5A via miR-370-3p/miR-136-5p.
Song D, Ye Z, Chen F, Zhan L, Sun X. Song D, et al. BMC Cancer. 2023 Sep 4;23(1):821. doi: 10.1186/s12885-023-11314-2. BMC Cancer. 2023. PMID: 37667251 Free PMC article.
References
- Ferlay J., et al. , Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). - PubMed
- Magee P. N., The experimental basis for the role of nitroso compounds in human cancer. Cancer Surv. 8, 207–239 (1989). - PubMed
- Yang C. S., Research on esophageal cancer in China: A review. Cancer Res. 40, 2633–2644 (1980). - PubMed
- Abnet C. C., et al. , Zinc concentration in esophageal biopsy specimens measured by X-ray fluorescence and esophageal cancer risk. J. Natl. Cancer Inst. 97, 301–306 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases