Nucleolar localization of the Notch4 intracellular domain underpins its regulation of the cellular response to genotoxic stressors - PubMed (original) (raw)
Nucleolar localization of the Notch4 intracellular domain underpins its regulation of the cellular response to genotoxic stressors
Neetu Saini et al. Cell Death Discov. 2020.
Abstract
Cell survival is one of the many cellular processes regulated by Notch family of proteins. A comparison of human breast cancer cell lines, which differ in the levels of endogenous Notch4, implicated the protein in regulating susceptibility to apoptosis triggered by genomic damage. In agreement with this observation, increased susceptibility to genotoxic damage was observed following siRNA ablations of Notch4 in two breast cancer cell lines. Further, overexpressing Notch4 intracellular domain (NIC4) tagged to GFP (NIC4-GFP), protected cells from apoptosis triggered by genotoxic drugs. In cells immune-stained for endogenous Notch4, protein was detected in the nucleolus and nucleoplasm, which was also confirmed by the co-localization of NIC4-GFP with RFP-tagged nucleolar proteins in breast cancer cells or the unrelated HEK cell line. Linking functional outcomes to nucleolar localization, NIC4-GFP protection from apoptosis, required the nucleolar proteins Nucleolin and Fibrillarin. Consistently, immunoprecipitation analysis revealed associations between nucleolar proteins-Nucleolin and Nucleophosmin-and Notch4. Microscopy-based biophysical analysis of live cells showed that nucleolar and nucleoplasmic pools of NIC4-GFP are mobile, with some sequestration of nucleolar NIC4-GFP pools. A nucleolar excluded form, NIC4_3RA-GFP, generated by site-directed mutagenesis of the nucleolar localization sequence in NIC4, could not protect from apoptosis triggered by genotoxic stressors. However, transcriptional activity or protection from apoptosis triggered by endoplasmic stress was comparable in cells expressing NIC4_3RA-GFP or NIC4-GFP. Together, the data show that nucleolar localization of NIC4 is critical for the regulation of genomic damage and may be uncoupled from its activities in the nucleoplasm. This study identifies intrinsic features of NIC4 that regulate signaling outcomes activated by the receptor by controlling its spatial localization.
Keywords: Apoptosis; Breast cancer.
© The Author(s) 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures
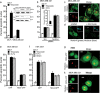
Fig. 1. Notch4 protects from apoptosis triggered by genotoxic agents.
a Apoptotic nuclear damage in MCF-7 and MDA-MB-231 cell lines continued untreated or treated with 10 μM etoposide for 24 h in serum-free medium. Inset: immunoblot of cell lysates probed for Notch4 and Notch1 proteins. b Apoptotic damage in MDA-MB-231 cells pretreated with siRNA to Notch4 or a scrambled control for 48 h and then continued untreated (UT) or treated with etoposide (10 μM) or 5-FU (10 μM) for 24 h in serum-free medium. Inset: percentage of Notch4 transcripts in the siRNA-treated groups. c Representative confocal images of cells stained for Notch4 (green) and Nucleolin (red) as described in “Methods” (Manders correlation coefficient: 0.73 ± 0.23). d Representative confocal images of cells stained for Notch4 in cells pretreated with the vehicle or GSI-X (10 μM) for 24 h in serum-free medium. e Apoptotic nuclear damage in cells expressing GFP or NIC4-GFP and treated with etoposide or 5-FU for 48 h in serum-free medium. f Percentage of apoptotic nuclear damage in HEK cells expressing GFP or NIC4-GFP and treated with etoposide or 5-FU or 4NQO (5 μM) for 48 h in serum-free medium. g Representative confocal images of HEK cells co-expressing NIC4-GFP and Fibrillarin-RFP imaged 24 h after transfection (Manders correlation coefficient: 0.87 ± 0.15). h Representative confocal images of MDA-MB-231 cells co-expressing NIC4-GFP and FBL-RFP imaged 24 h after transfection (Manders correlation coefficient: 0.96 ± 0.07). Apoptotic nuclear damage was scored by visualizing nuclei stained with Hoechst 33342. Data represent the mean ± S.D. of three independent experiments. Scale bar: 5 μm.
Fig. 2. Nucleolar proteins and the DNA damage response proteins are required for NIC4-mediated anti-apoptotic activity.
a–f Apoptotic nuclear damage in HEK cells pretreated with siRNA shown in each panel, transfected with plasmids as shown and treated with 10 μM etoposide or 10 μM 5-FU or 5 μM 4NQO for 48 h in serum-free medium. Insets: percentage of mRNA levels in the transfected groups. Data are mean ± S.D. of three independent experiments.
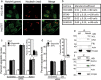
Fig. 3. Notch4 localization in breast cancer cell lines.
a Representative confocal images of cell lines stained for Notch4 (green) or Nucleolin (red) and merged images. b Co-localization of Nucleolin (red) and Notch4 (green), quantified by Manders Coefficient (described in “Methods”) for the indicated cell lines. c, d Apoptotic nuclear damage in Hs578T cells pretreated with the indicated siRNA for 48 h and then cultured untreated (UT) or with 10 μM etoposide or 10 μM 5-FU for 24 h. Inset: percentage of mRNA levels in siRNA-transfected groups. e, f MDA-MB-231 cells treated with etoposide for 6 h were lysed and subjected to immunoprecipitation with antibody to Notch4 (e) or NCL (f) and IgG (isotype control). Immunoprecipitates were analyzed by western blotting for NCL, Notch4, NPM, FBL, Actin, and IgG. Data show the mean ± S.D. of three independent experiments. Scale bar: 5 μm.
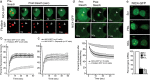
Fig. 4. Cellular dynamics of NIC4-GFP.
a Representative confocal images of a cell co-expressing FBL-RFP and NIC4-GFP. The white arrowhead indicates a bleached nucleolus. b, c Fluorescence intensity recovered over time following photo-bleaching of cells co-expressing FBL-RFP and NIC4-GFP (b) and treated for 6 h with 10 μM etoposide (c). Data plotted are mean ± SD of 20 cells in b and 37 cells in c. d Loss of fluorescence intensity over time (FLIP) in cells co-expressing FBL-RFP and NIC4-GFP. The panel shows a cell over time, with the visualization spot (not bleached) marked by an arrow-head, surrounded by the bleached (dark) region. Change in fluorescence over time is plotted in the graph below. Data plotted are mean ± S.D. of a minimum of 19 cells in each condition. e Representative confocal images of HEK cells expressing NIC4-GFP following treatment with siRNA shown. mRNA levels of the indicated transcripts in siRNA-treated cells is plotted. Images are representative of 30 cells in each condition. Scale bar: 5 μm.
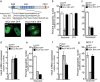
Fig. 5. Nucleolar localization of NIC4 is required for protection from genomic stress.
a Schematic showing the putative NoLS sequence in NIC4, the modifications in NIC4_3RA (with three R residues replaced by A in the NoLS), and the additional NoLS at the NIC4 N-terminal. Representative confocal images of HEK cells expressing NIC4_3RA-GFP (left) and NoLS_NIC4 GFP (right). Scale bar: 5 μm. b, c Apoptotic nuclear damage in cells expressing GFP, NIC4-GFP, or NIC4_3RA-GFP, treated with 10 μM etoposide or 10 μM 5-FU for 48 h (b) or 10 μM Thapsigargin for 20 h (c). d Relative transcript levels of Hes5 in cells transfected with GFP, NIC4-GFP, or NIC4_3RA-GFP and cultured for 36 h in complete medium. e Apoptotic nuclear damage in cells expressing GFP, NIC4-GFP, or NoLS_NIC4-GFP, treated with etoposide for 48 h in serum-free medium. f Relative transcript levels of Hes5 in cells transfected with the indicated plasmids and cultured for 36 h. g Apoptotic nuclear damage in MCF7 cells expressing GFP, NIC4-GFP, or NoLS_NIC4-GFP, treated with etoposide for 48 h in serum-free medium. Data plotted are mean ± S.D. of three independent experiments.
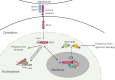
Fig. 6. Schematic summarizing key observations of the current study.
NIC4 mobility in the nucleus and nucleolus is dynamic with NIC4 localization to the nucleolus, guided by its NoLS. NIC4 associates with NCL and NPM, and protection from genomic stressors is dependent on nucleolar proteins NCL and FBL. Nucleolar functions can be uncoupled from nuclear activities of NIC4. Not to scale.
Similar articles
- Sirtuin1 meditated modification of Notch1 intracellular domain regulates nucleolar localization and activation of distinct signaling cascades.
Saini N, Bheeshmachar G, Sarin A. Saini N, et al. Front Cell Dev Biol. 2022 Sep 23;10:988816. doi: 10.3389/fcell.2022.988816. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36211456 Free PMC article. - Immunolocalization of nucleolar proteins during bovine oocyte growth, meiotic maturation, and fertilization.
Fair T, Hyttel P, Lonergan P, Boland MP. Fair T, et al. Biol Reprod. 2001 May;64(5):1516-25. doi: 10.1095/biolreprod64.5.1516. Biol Reprod. 2001. PMID: 11319160 - Notch4 inhibits endothelial apoptosis via RBP-Jkappa-dependent and -independent pathways.
MacKenzie F, Duriez P, Wong F, Noseda M, Karsan A. MacKenzie F, et al. J Biol Chem. 2004 Mar 19;279(12):11657-63. doi: 10.1074/jbc.M312102200. Epub 2003 Dec 29. J Biol Chem. 2004. PMID: 14701863 - Nucleolin and nucleophosmin: nucleolar proteins with multiple functions in DNA repair.
Scott DD, Oeffinger M. Scott DD, et al. Biochem Cell Biol. 2016 Oct;94(5):419-432. doi: 10.1139/bcb-2016-0068. Epub 2016 Jun 29. Biochem Cell Biol. 2016. PMID: 27673355 Review. - Nucleolar stress: Friend or foe in cardiac function?
Yan D, Hua L. Yan D, et al. Front Cardiovasc Med. 2022 Oct 31;9:1045455. doi: 10.3389/fcvm.2022.1045455. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36386352 Free PMC article. Review.
Cited by
- Targeting Notch4 in Cancer: Molecular Mechanisms and Therapeutic Perspectives.
Xiu M, Zeng X, Shan R, Wen W, Li J, Wan R. Xiu M, et al. Cancer Manag Res. 2021 Sep 8;13:7033-7045. doi: 10.2147/CMAR.S315511. eCollection 2021. Cancer Manag Res. 2021. PMID: 34526819 Free PMC article. Review. - Sirtuin1 meditated modification of Notch1 intracellular domain regulates nucleolar localization and activation of distinct signaling cascades.
Saini N, Bheeshmachar G, Sarin A. Saini N, et al. Front Cell Dev Biol. 2022 Sep 23;10:988816. doi: 10.3389/fcell.2022.988816. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36211456 Free PMC article. - The Clinical Application of Immunohistochemical Expression of Notch4 Protein in Patients with Colon Adenocarcinoma.
Brzozowa-Zasada M, Piecuch A, Michalski M, Matysiak N, Kucharzewski M, Łos MJ. Brzozowa-Zasada M, et al. Int J Mol Sci. 2023 Apr 19;24(8):7502. doi: 10.3390/ijms24087502. Int J Mol Sci. 2023. PMID: 37108670 Free PMC article. - Novel INHAT repressor drives glioblastoma growth by promoting ribosomal DNA transcription in glioma stem cells.
Tao W, Lei H, Luo W, Huang Z, Ling P, Guo M, Wan L, Zhai K, Huang Q, Wu Q, Xu S, Zeng L, Wang X, Dong Z, Rich JN, Bao S. Tao W, et al. Neuro Oncol. 2023 Aug 3;25(8):1428-1440. doi: 10.1093/neuonc/noac272. Neuro Oncol. 2023. PMID: 36521011 Free PMC article. - Notch1 Modulation of Cellular Calcium Regulates Mitochondrial Metabolism and Anti-Apoptotic Activity in T-Regulatory Cells.
Saini N, Lakshminarayanan S, Kundu P, Sarin A. Saini N, et al. Front Immunol. 2022 Feb 10;13:832159. doi: 10.3389/fimmu.2022.832159. eCollection 2022. Front Immunol. 2022. PMID: 35222416 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous