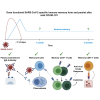
Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19 - PubMed (original) (raw)
. 2021 Jan 7;184(1):169-183.e17.
doi: 10.1016/j.cell.2020.11.029. Epub 2020 Nov 23.
Jason Netland 1, Laila Shehata 1, Kurt B Pruner 1, Peter A Morawski 2, Christopher D Thouvenel 3, Kennidy K Takehara 1, Julie Eggenberger 4, Emily A Hemann 4, Hayley R Waterman 2, Mitchell L Fahning 2, Yu Chen 3, Malika Hale 3, Jennifer Rathe 4, Caleb Stokes 4, Samuel Wrenn 5, Brooke Fiala 5, Lauren Carter 5, Jessica A Hamerman 6, Neil P King 5, Michael Gale Jr 4, Daniel J Campbell 6, David J Rawlings 7, Marion Pepper 8
Affiliations
- PMID: 33296701
- PMCID: PMC7682481
- DOI: 10.1016/j.cell.2020.11.029
Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19
Lauren B Rodda et al. Cell. 2021.
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus is causing a global pandemic, and cases continue to rise. Most infected individuals experience mildly symptomatic coronavirus disease 2019 (COVID-19), but it is unknown whether this can induce persistent immune memory that could contribute to immunity. We performed a longitudinal assessment of individuals recovered from mild COVID-19 to determine whether they develop and sustain multifaceted SARS-CoV-2-specific immunological memory. Recovered individuals developed SARS-CoV-2-specific immunoglobulin (IgG) antibodies, neutralizing plasma, and memory B and memory T cells that persisted for at least 3 months. Our data further reveal that SARS-CoV-2-specific IgG memory B cells increased over time. Additionally, SARS-CoV-2-specific memory lymphocytes exhibited characteristics associated with potent antiviral function: memory T cells secreted cytokines and expanded upon antigen re-encounter, whereas memory B cells expressed receptors capable of neutralizing virus when expressed as monoclonal antibodies. Therefore, mild COVID-19 elicits memory lymphocytes that persist and display functional hallmarks of antiviral immunity.
Keywords: COVID-19; SARS-CoV2; adaptive immune response; human; memory B cell; memory T cell; monoclonal antibody; vaccine.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests M.P., D.J.R., J.N., C.D.T., Y.C., and L.B.R. have filed a patent under the provisional serial no. 63/063,841. Other authors declare no competing interests.
Figures
Graphical abstract
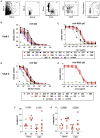
Figure 1
Mild COVID-19 Induces Persistent, Neutralizing Anti-SARS-CoV-2 IgG Antibody (A) Study timeline. Range is indicated by box and median indicated by line for each event. (B) ELISA dilution curves and AUC for anti-RBD IgG (left), IgM (center), and IgA (right) from HC and CoV2+ plasma samples at Visit 1 (V1) and Visit 2 (V2). Dashed line indicates mean + 3 SD of the HC AUC values. (C) Comparing V1 and V2 AUC in HC and CoV2+ individuals for each antibody isotype. V2 AUC values were normalized to V1 samples run with V2 samples. (D) Percent inhibition of RBD binding to ACE2 by plasma sVNT at 1:2 plasma dilution. (E) Spearman correlation between percent RBD inhibition by sVNT at a 1:2 plasma dilution and anti-RBD IgG AUC at both visits. (F) Percent RBD inhibition at 1:2 plasma dilution at V1 and V2, paired by sample. (G) Spearman correlation between percent RBD inhibition by sVNT at a 1:2 plasma dilution and percent virus neutralization by PRNT at a 1:160 plasma dilution. (H) CoV2+ percent virus neutralization by PRNT at a 1:160 plasma dilution normalized and paired as in (C). Statistics for unpaired data determined by 2-tailed Mann-Whitney tests and, for paired data, by 2-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. See also Figures S1 and S4.
Figure S1
Healthy Controls Do Not Have SARS-CoV-2 RBD or Spike-Specific Antibodies, Related to Figure 1 ELISA dilution curves and area under the curve (AUC) for anti-RBD and anti-spike IgG (left) and IgM (right) in plasma collected from individuals prior to 2020 and the SARS-CoV-2 pandemic (historical negatives, HN, black), from healthy controls (HC, at Visit 2) and from individuals that tested PCR+ for SARS-CoV-2 (CoV2+, at Visit 1). Dashed line indicates mean + 3 SD of HN AUC values. Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD.
Figure S2
PBMC Innate Populations in CoV2+ Individuals Return to Immune Quiescence by Visit 1, Related to Figure 1 (A) Flow cytometry gating for CD15-CD3-CD19-CD56-HLADR+CD14+ monocytes (purple gate), which were further divided into CD14loCD16+ (red gate), CD14+CD16+ (blue gate), and CD14+CD16- monocytes (green gate), and CD15-CD3-CD19-CD56-CD14-CD304+CD123+ plasmacytoid dendritic cells (pDCs) (pink gate). (B) Percent monocytes and pDCs of live PBMCs from healthy controls (HC) and previously SARS-CoV-2 infected (CoV2+) individuals. (C) Percent subsets of monocytes from PBMCs. Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from two experiments.
Figure S3
Bulk PBMCs Return to Immune Quiescence by Visit 1, Related to Figure 1 (A and B) Representative flow cytometry plots and frequencies of αβ and γδ T cell subsets at Visit 1 (V1) in PBMCs from healthy control (HC) and SARS-CoV-2-recovered (CoV2+) individuals. (C and D) Representative flow cytometry plots and frequencies of CD4+ and CD8+ T cell effector/activation states (Ki67+, T-bet+, HLA-DR+CD38+) of total non-naive, memory CD45RA+CCR7+/− CD4+ or CD8+ T cells at V1 in HC and CoV2+ PBMCs. (E and F) Representative flow cytometry plots and frequencies of CD4+ memory and T-helper subsets at V1 in HC and CoV2+ PBMCs. (G and H) Representative flow cytometry plots and frequencies of cTfh (CXCR5+CD45RA-) and cTfh activation (ICOS+PD-1+) and helper (CXCR3+/−CCR6+/−) subsets at V1 in HC and CoV2+ PBMCs. (I) Frequency of B cells (CD19+CD3-) at V1 in HC and CoV2+ PBMCs. Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from two experiments.
Figure S4
Mild COVID-19 Induces Persistent, Neutralizing Anti-SARS-CoV-2 IgG Antibody, Related to Figure 1 (A) ELISA dilution curves and area under the curve (AUC) for anti-spike IgG (left), IgM (center), and IgA (right) from healthy control (HC) and SARS-CoV-2-recovered (CoV2+) individuals plasma at Visit 1 (V1). Dashed line indicates mean + 3 SD of the HC AUC values. (B) Spearman correlation of V1 anti-RBD and anti-spike IgG (left), IgM (center), and IgA (right) AUC. Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD.
Figure S5
Mild COVID-19 Induces a Sustained Enrichment of RBD-Specific IgG+ Memory B Cells, Related to Figure 2 (A) Representative flow cytometry gates for phenotyping RBD-specific B cells from PBMCs in Figure 2 set on total B cells from a healthy control (HC) (surface stain, top; intracellular stain, bottom). (B) Number of RBD-specific IgD+, IgM+ and IgA+ MBCs (CD20+RBD tetramer+decoy tetramer- CD27+CD21+/CD27+CD21-/CD27-CD21-) from healthy control (HC) and SARS-CoV-2-recovered (CoV2+) PBMCs at Visit 1 (V1) and Visit 2 (V2). Statistics for unpaired data determined by two-tailed Mann-Whitney tests and, for paired data, by two-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.02. Error bars represent mean and SD. Data from two experiments per visit.
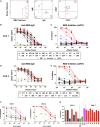
Figure 2
Mild COVID-19 Induces a Sustained Enrichment of RBD-Specific IgG+ Memory B Cells (A) Representative gating of live CD3–CD14–CD16– cells for SARS-CoV-2 RBD-specific cells (RBD tetramer+decoy tetramer–) from CoV2+ and HC PBMCs at V1 and V2. (B) Number of RBD-specific B cells (RBD tetramer+decoy tetramer–CD20+) per 1 × 106 PBMCs. (C) Representative flow cytometry plots and number of RBD-specific PBs (RBD tetramer+decoy tetramer−CD20−CD138hi) (na = could not be calculated because all values 0). (D) Representative gating of RBD-specific B cells for naive B cells (CD21+CD27−) and MBCs (CD21+CD27+/CD21–CD27+/CD21–CD27– populations outlined in green). (E) Proportion of RBD-specific B cells that are naive (CD21+CD27−), classical MBCs (CD21+CD27+), or activated MBCs (CD21−CD27+/−), statistics for the proportion that are MBCs. (F) Number of RBD-specific MBCs (classical and activated). (G) Representative gating of RBD-specific MBCs for BCR isotype (IgD, IgM, IgA, and IgG) expression. (H) Proportion of RBD-specific MBCs expressing the BCR isotypes IgD, IgM, IgA, and IgG. Statistics are for the proportion that are IgG+. (I) Number of RBD-specific IgG+ MBCs. (J) Representative gating of RBD-specific MBCs for T-bet expression and number of RBD-specific T-bet+ MBCs. Statistics for unpaired data determined by 2-tailed Mann-Whitney tests and, for paired data, by 2-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.04. Error bars represent mean and SD. Data from 2 experiments per visit. See also Figure S5.
Figure 3
SARS-CoV-2 Infection Induces Durable, Functional Spike-Reactive CD4+ T Cells (A) Representative flow cytometry plots of ICOS and CD40L expression on antigen-experienced (non-CD45RA+CCR7+) CD4+ T cells 20 h after incubation of PBMCs from HC and CoV2+ individuals at V1 and V2 with vehicle or SARS-CoV-2 spike. (B) Number of antigen-experienced ICOS+CD40L+CD4+ T cells per 1x106 CD4+ T cells from HC and CoV2+ samples after incubation with vehicle (Veh.) or spike (S) at both time points (right) and calculated number of spike-responsive CD4+ T cells (number after incubation with spike minus number after incubation with vehicle) compared across time points (left). (C) Number of antigen-experienced CXCR5+ICOS+CD40L+CD4+ T cells (cTfh) per 1 × 106 CD4+ T cells from HC and CoV2+ samples after incubation with Veh. or S at both time points (right) and calculated number of spike-responsive cTfh cells (number after incubation with spike minus number after incubation with vehicle) compared across time points (left). (D) Representative flow cytometry plots of sorted CD4+ naive (CD45RA+CCR7+), TCM (CD45RA−CCR7+), or TEM (CD45RA−CCR7−) T cells from HC and CoV2+ PBMCs after 5–6 days of co-culture with SARS-CoV-2 spike-protein-pulsed autologous monocytes and measuring proliferation by CPD dilution. (E) SARS-CoV-2 spike-specific expansion of sorted CD4+ naive T, TCM, and TEM cells from V1 (circles) and V2 (squares) reported as frequency of CXCR3+CPDlo cells after incubation with spike minus frequency after incubation with vehicle. (F) Number of cytokine-producing, antigen-experienced CD69+CD4+ T cells per 1 × 106 CD4+ T cells after incubation with Veh. or S (right) and calculated number of spike-responsive, cytokine-producing CD4+ T cells (number after incubation with spike minus number after incubation with vehicle) (left). (G) Frequency of antigen-experienced CD69+CD4+ T cell subsets, CCR6+/− Teff (CXCR5−), and CCR6+/− cTfh (CXCR5+) producing IL-2, IFN-γ, and IL-17A effector cytokines after incubation with spike for 20 h. (H) Representative flow cytometry plots of CD69 and IFN-γ expression on antigen-experienced CD8+ T cells from HC and CoV2+ PBMCs at V2 after 20 h of incubation with vehicle or SARS-CoV-2 spike. (I) Number of antigen-experienced IFN-γ+CD69+CD8+ T cells per 1 × 106 CD8+ T cells after 20 h of incubation with Veh. or S (right) and calculated number of spike-responsive CD8+ T cells (number after incubation with spike minus number after incubation with vehicle) (left). Statistics for unpaired data determined by 2-tailed Mann-Whitney tests and, for paired data, by 2-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from 2 experiments per visit. See also Figure S6.
Figure S6
SARS-CoV-2 Infection Induces Durable, Functional Spike-Reactive CD4+ T Cells, Related to Figure 3 (A) Flow cytometry sorting strategy for naive, T central memory (TCM), and T effector memory (TEM) cells from HC and CoV2+ PBMCs at Visit 1 and Visit 2 before 5-6 days of culture with autologous monocytes and SARS-CoV-2 spike protein or vehicle. (B) Representative flow cytometry gating on PMA/Ionomycin-activated PBMCs for cytokine expression by antigen experienced (non-CD45RA+CCR7+) CD4+ T cells subset into CCR6+/− T effector cells (Teff, CXCR5-) and circulating T follicular helper cells (cTfh, CXCR5+). (C) Representative flow cytometry gating on antigen-experienced (non-CD45RA+CCR7+) CD4+ T cells from HC and CoV2+ V2 PBMCs following incubation with SARS-CoV-2 spike for 20 h. Gating on CD69+ CCR6+/− T effector cells (Teff, CXCR5-) and CCR6+/− circulating T follicular helper cells (cTfh, CXCR5+) for IL-2, IFN-γ and IL-17A effector cytokines expression. (D) Number of IL-4-producing, antigen-experienced CD69+CD4+ T cells per 1x106 CD4+ T cells after incubation with vehicle (Veh.) or SARS-CoV2 spike (S) (left) and calculated number of spike-responsive, cytokine-producing CD4+ T cells (number after incubation with spike minus number after incubation with vehicle)(right). Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from two experiments per visit.
Figure S7
SARS-CoV-2-Specific MBCs Can Express Neutralizing Antibodies, Related to Figure 4 and Table S1 (A) Gating strategy for sorting RBD-specific B cells. (B) IgG ELISA to confirm expression of Visit 1 antibodies in transfected cell culture supernatants. Positive control is the kit standard (std) and negative control is supernatant from untransfected cells (no trans, green). (C) RBD ELISA of purified Visit 1 monoclonal antibodies. Negative control (green) is an irrelevant _Plasmodium_-specific antibody. (D) IgG ELISA to confirm expression of Visit 2 antibodies in transfected cell culture supernatants. (E) RBD ELISA of purified Visit 2 monoclonal antibodies. (F) Number of mutations in variable regions of RBD-specific monoclonal antibodies. (G) Mutation frequency of variable regions of RBD-specific monoclonal antibodies. Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05.
Figure 4
SARS-CoV-2-Specific MBCs Can Express Neutralizing Antibodies (A) Representative flow plots of index-sorted RBD-tetramer specific B cells (gating scheme in Figure S7A). BCRs cloned from cells are shown in red. (B) Anti-RBD ELISA of culture supernatants from cells transfected to express one of the Visit 1 monoclonal antibodies or supernatant from untransfected cells (no trans). Antibodies that did not bind RBD are shown in orange. (C) Inhibition of RBD binding to ACE2 by Visit 1 monoclonal antibody supernatants measured by sVNT assay, compared to a known RBD-specific neutralizing antibody (Ty1). Red indicates strong inhibitors, blue moderate inhibitors and black non-inhibitors. (D) Anti-RBD ELISA of culture supernatants from cells transfected to express one of the Visit 2 monoclonal antibodies. Antibodies that did not bind RBD are shown in orange. (E) Inhibition of RBD binding to ACE2 by Visit 2 monoclonal antibodies measured by sVNT assay. Red indicates strong inhibitors and black non-inhibitors. (F) Neutralization capacity of purified monoclonal antibodies as measured by PRNT. 2B04 and 2C02 are previously identified strong and weak neutralizing murine antibodies, respectively, and MSP-003 is an irrelevant _Plasmodium_-specific antibody. (G) IC50 values calculated from PRNT. Dotted line represents the limit of detection. See also Figure S7.
Figure 5
Recovered Individuals Formed Multifaceted SARS-CoV-2-Specific Immune Memory Heatmap of values for independent SARS-CoV2 RBD- or spike-specific immune memory components from each HC and CoV2+ individual at Visit 2. RBD-specific IgG measured by ELISA (AUC). Percent inhibition by sVNT calculated at 1:2 plasma dilution. Number of RBD-specific IgG+ MBCs per 1 × 106 PBMCs. Number of spike-responsive (CD69+), cytokine-producing (IL-2/IFN-γ/IL-17A), antigen-experienced CD4+ T cells calculated by number after 20 h incubation with spike minus number after incubation with vehicle. Number of spike-responsive (CD69+IFN-γ+), antigen-experienced CD8+ T cells calculated by number after 20 h incubation with spike minus number after incubation with vehicle. The color scales are set for each metric (row) with the mean + 1 SD of the HC set to white.
Update of
- Functional SARS-CoV-2-specific immune memory persists after mild COVID-19.
Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PM, Thouvenel C, Takehara KK, Eggenberger J, Hemann EA, Waterman HR, Fahning ML, Chen Y, Rathe J, Stokes C, Wrenn S, Fiala B, Carter LP, Hamerman JA, King NP, Gale M, Campbell DJ, Rawlings D, Pepper M. Rodda LB, et al. medRxiv [Preprint]. 2020 Aug 15:2020.08.11.20171843. doi: 10.1101/2020.08.11.20171843. medRxiv. 2020. PMID: 32817957 Free PMC article. Updated. Preprint. - Functional SARS-CoV-2-specific immune memory persists after mild COVID-19.
Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel C, Takehara KK, Eggenberger J, Hemann E, Waterman HR, Fahning ML, Chen Y, Rathe J, Stokes C, Wrenn S, Fiala B, Carter L, Hamerman JA, King NP, Gale M Jr, Campbell DJ, Rawlings D, Pepper M. Rodda LB, et al. Res Sq [Preprint]. 2020 Aug 13:rs.3.rs-57112. doi: 10.21203/rs.3.rs-57112/v1. Res Sq. 2020. PMID: 32818218 Free PMC article. Updated. Preprint.
Similar articles
- Functional SARS-CoV-2-specific immune memory persists after mild COVID-19.
Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel C, Takehara KK, Eggenberger J, Hemann E, Waterman HR, Fahning ML, Chen Y, Rathe J, Stokes C, Wrenn S, Fiala B, Carter L, Hamerman JA, King NP, Gale M Jr, Campbell DJ, Rawlings D, Pepper M. Rodda LB, et al. Res Sq [Preprint]. 2020 Aug 13:rs.3.rs-57112. doi: 10.21203/rs.3.rs-57112/v1. Res Sq. 2020. PMID: 32818218 Free PMC article. Updated. Preprint. - Functional SARS-CoV-2-specific immune memory persists after mild COVID-19.
Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PM, Thouvenel C, Takehara KK, Eggenberger J, Hemann EA, Waterman HR, Fahning ML, Chen Y, Rathe J, Stokes C, Wrenn S, Fiala B, Carter LP, Hamerman JA, King NP, Gale M, Campbell DJ, Rawlings D, Pepper M. Rodda LB, et al. medRxiv [Preprint]. 2020 Aug 15:2020.08.11.20171843. doi: 10.1101/2020.08.11.20171843. medRxiv. 2020. PMID: 32817957 Free PMC article. Updated. Preprint. - Persistence of Antibody and Cellular Immune Responses in Coronavirus Disease 2019 Patients Over Nine Months After Infection.
Yao L, Wang GL, Shen Y, Wang ZY, Zhan BD, Duan LJ, Lu B, Shi C, Gao YM, Peng HH, Wang GQ, Wang DM, Jiang MD, Cao GP, Ma MJ. Yao L, et al. J Infect Dis. 2021 Aug 16;224(4):586-594. doi: 10.1093/infdis/jiab255. J Infect Dis. 2021. PMID: 33978754 Free PMC article. - Infection and Immune Memory: Variables in Robust Protection by Vaccines Against SARS-CoV-2.
Ahluwalia P, Vaibhav K, Ahluwalia M, Mondal AK, Sahajpal N, Rojiani AM, Kolhe R. Ahluwalia P, et al. Front Immunol. 2021 May 11;12:660019. doi: 10.3389/fimmu.2021.660019. eCollection 2021. Front Immunol. 2021. PMID: 34046033 Free PMC article. Review. - Humoral Immunity against SARS-CoV-2 and the Impact on COVID-19 Pathogenesis.
Lee E, Oh JE. Lee E, et al. Mol Cells. 2021 Jun 30;44(6):392-400. doi: 10.14348/molcells.2021.0075. Mol Cells. 2021. PMID: 34059562 Free PMC article. Review.
Cited by
- Identification of virus epitopes and reactive T-cell receptors from memory T cells without peptide synthesis.
Wang L, Xu R, Huang D, Peng P, Sun K, Hu J, Liu BZ, Fang L, Zhang L, Sun X, Gu F, Tang N, Huang AL, Lin X, Lan X. Wang L, et al. Commun Biol. 2024 Nov 4;7(1):1432. doi: 10.1038/s42003-024-07048-x. Commun Biol. 2024. PMID: 39496850 Free PMC article. - Antibody Levels Poorly Reflect on the Frequency of Memory B Cells Generated following SARS-CoV-2, Seasonal Influenza, or EBV Infection.
Wolf C, Köppert S, Becza N, Kuerten S, Kirchenbaum GA, Lehmann PV. Wolf C, et al. Cells. 2022 Nov 18;11(22):3662. doi: 10.3390/cells11223662. Cells. 2022. PMID: 36429090 Free PMC article. - Immune profiling of COVID-19: preliminary findings and implications for the pandemic.
Maecker HT. Maecker HT. J Immunother Cancer. 2021 May;9(5):e002550. doi: 10.1136/jitc-2021-002550. J Immunother Cancer. 2021. PMID: 33963016 Free PMC article. Review. - Atypical and non-classical CD45RBlo memory B cells are the majority of circulating SARS-CoV-2 specific B cells following mRNA vaccination or COVID-19.
Priest DG, Ebihara T, Tulyeu J, Søndergaard JN, Sakakibara S, Sugihara F, Nakao S, Togami Y, Yoshimura J, Ito H, Onishi S, Muratsu A, Mitsuyama Y, Ogura H, Oda J, Okusaki D, Matsumoto H, Wing JB. Priest DG, et al. Nat Commun. 2024 Aug 9;15(1):6811. doi: 10.1038/s41467-024-50997-4. Nat Commun. 2024. PMID: 39122676 Free PMC article. - Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection.
Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP. Khoury DS, et al. Nat Med. 2021 Jul;27(7):1205-1211. doi: 10.1038/s41591-021-01377-8. Epub 2021 May 17. Nat Med. 2021. PMID: 34002089
References
- Ahmed R., Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. - PubMed
- Alamyar E., et al. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods in molecular biology (Clifton, N.J.) 882(Chapter. 2012;32:569–604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI118803/AI/NIAID NIH HHS/United States
- R21 AI158788/AI/NIAID NIH HHS/United States
- T32 AI106677/AI/NIAID NIH HHS/United States
- S10 OD024979/OD/NIH HHS/United States
- P30 DK089507/DK/NIDDK NIH HHS/United States
- T32 AI007140/AI/NIAID NIH HHS/United States
- R01 AI127726/AI/NIAID NIH HHS/United States
- R01 AI150178/AI/NIAID NIH HHS/United States
- TL1 TR002318/TR/NCATS NIH HHS/United States
- U01 AI142001/AI/NIAID NIH HHS/United States
- U01 AI151698/AI/NIAID NIH HHS/United States
- U19 AI125378/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous